Isolation and Genetic Identification of L-Asparaginase Antitumor from Streptomyces sp.
Received: 29 July 2019 / Received in revised form: 17 January 2020, Accepted: 24 January 2020, Published online: 28 February 2020
© Biochemical Technology Society 2014-2020
© Sevas Educational Society 2008
Abstract
L-asparaginase (L-asparagine amidohydrolase, E.C.3.5.1.1) is a supplementary cellular enzyme used as an anticancer substance. 31 Streptomyces strains isolated from rhizosphere of some plants in different regions of Saudi Arabia. The highest 11 Streptomyces isolates in L-asparaginase activity were selected for further molecular analysis based on differences in 16S rRNA gene sequence. Alignment analysis using the BLAST tools confirmed that all isolates are related to Streptomyces with a similarity of 97% to 100%. And partial-length gene sequence was sentinto NCBI Gen Bank and received the accession numbers (Streptomyces griseoruben MN588164, Streptomyces alboflavus MN588165, Streptomyces werraensis MN588166, Streptomyces spororaveus MN588167, Streptomyces chryseus MN588168, Streptomyces globosus MN588169, Streptomyces roseolilacinus MN588170, Streptomyces xanthophaeus MN588171, Streptomyces lavendulae MN588172, Streptomyces tuirus MN588174, and Streptomyces tendae MN588175).
Key words: Anticancer, L-asparaginase, Streptomyces
Introduction
L-asparaginase is the hydrolyzate linkage of amide in L-asparagine to L-aspartic acid and ammonia (Kumar and Verma, 2012) and has been thoroughly investigated as the first enzyme with anti-leukemic activity (Savitri and Azmi, 2003). In chemotherapy for acute lymphoblastic leukemia (ALL), antinoplastic agent is used (Narta et al., 2007).
The increased attantion to L-asparaginase was related to many of the features used in potential industrial fields (Pedreschi et al., 2008) and was modified as anti-leukemia therapy in lymphoblastic leukemia (Patil et al., 2011; and Pieters et al., 2011 and Jain et al., 2012). Erwinia chrysanthemi and E. coli have been used as effective drugs for lymphoblastic leukemia a (Kozak et al., 2002; and Graham, 2003) without toxicity (Duval et al., 2002).
Streptomyces found in marine and terrestrial reigns (Pathom-Aree et al., 2006) and produce 50% of the discovered bioactive secondary metabolites (Manivasagan et al., 2013). L-asparaginase can be produced from Streptomyces sp. (Basha et al., 2009), and Streptomyces gulbargensis (Amena et al., 2010). Streptomyces noursei MTCC 10469 isolated from the marine sponge Callyspongia diffusa (Dharmaraj, 2011).
Streptomyces aurantiacus (Gupta et al., 2007), which were classified in several ways as ribosomal proteins (Ochi, 1995), fatty acid (Zhang et al., 2007) and the 16S rRNA gene (Ramos et al. 1997; Prasad et al. 2013; Radhakrishnan et al., 2013). Moerever, 16S rRNA is suitable for phylogenetic analysis (Song et al., 2001).
Materials and Methods:
Considering the importance of soil properties, Sixty-one soil samples from plant rhizosphere were collected in Jeddah city, from costal soil and from algae in Thual. In addition, .some samples of cultivated soils in Al-Baha were also collected and studied. Soil samples collected from plants rhizosphere were 10 to 20 cm in depth and were stored at 4 ° C until use. (Alhaithloul H. A. S. 2019; Anisimova, 2019)
Streptomycesspp.Isolation:
All soil samples were placed in the oven to dry at a temperature 55° C to ensure the killing of the highest possible number of vegetative reproductions of bacterial species,, 90 ml of sterile dist. H2O from each sample was add to 10g, then serial dilution was prepared. Starch nitrate agar medium was inoculated for Streptomyces isolation (Waksman, 1962) with 0.1 ml of three different dilutions (10-4, 10-5 and 10-6). Three replicates were used for each dilution and the samples were incubated at 28± 2ºC for 7 days to give Streptomyces colonies.
Purification of isolated Streptomycesstrains:
Streptomyces colonies were characterized by their sharp edges, cretaceous form, and adhesion to the growth plate. Repeat sub-culturing for colonies to achieve pure culturing strains. Purified Streptomyces strains were isolated on 30% (v/v) glycerol at −20° C.
Detection of L-asparaginaseactivity from Streptomyces isolates (Dhanam and Kannan, 2015).
By rapid plate assay on M9 agar medium,an indicator was added to the media, pink colour zones around the colonies were reflected L-asparaginaseactivity (Rajguru and Deshmukh, 2016).
Quantitative of L-asparaginase activity Using the agar well Diffusion Technique (Rajguru and Deshmukh, 2016)
This technique was based on observations of greater enzyme production extracted from an extracellular source to cultures. Prepared plates contain M9 agar medium with red phenol.
The plates were pierced at diameter 8mm, then 100 µl of the crude enzyme concentrations were poured into the agar for each strain. At room temperature, leave the plates for 24 hours, and then observe diffuse into the medium by measuring the zone diameter (mm) of the L-asparaginase activity that appears as a pink-coloured area around the well.
Measurement of L-asparaginase activity using Nesslerization method:
Quantitative assay of L-asparaginase according to Mashburn, 1964. Measurement of ammonia release using the Nessler’s reaction. this mixture was incubated and after 10 min at 37° C 0.5 ml of 1.5 M Trichloroacetic acid (TCA). Precipitated protein was removed by centrifugation (10,000 rpm) for 5 min. 0.5 ml of diluted supernatant is dilutedto 7.0 ml. H2O and 1.0 ml of Nessler’s reagent were added and incubated at room temperature for 10 min. UV/Visible Spectrophotometer was used to measure the absorbance at 450 nm.
Molecular characterization of bacterial isolate.
using GeneJET (Thermo Fisher Scientific) Genomic DNA Purification Kit for Genomic DNA isolation.. By using the highly conserved global primers pA 5’AGA GTT TGA TCC TGG CTC AG 3’ and pH 5’ AAG GAG GTG ATC CAG CCG CA 3’, the amplification of the 1500 bp fragments representing the full length of the 16S rRNA gene was amplified (Edwards et al., 1989). 50 ng DNA, 1μL of each 10 μM primer, 12.5μL GoTaq® Green Master Mix (Promega, USA) and sterile dH2O up to 25 μl.
PCR amplification conditions:
Initial: incubation at 94 °C for 5 minutes, Denaturation: 94 °C for 1 min , Annealing: 55 °C for 1 min and Extension: 72 °C for 2 min , Number of cycles: 35. Final extension: 72 °C for 10 min.
Obtained nucleotide sequences of the genes 16S rRNAwere aligned using codon code aligner software (Barth et al., 2007). The BLAST algorithm and RDP database were used to analyzed and compared with those in GenBank to check for close evolutionary relatives.In order to characterize the strain, the nucleotide sequence of thegene of 16S rRNA was determined and the phylogenetic tree was constructed using the Neighbour- Joining (N-J) method Clustal-W in MegaAlign tool from DNASTAR version 12.3.1. DNA Star inc.
Final phylogenetic tree obtained using iTOL tree of life tool from Letunic and Bork (2019)
Results:
Isolation and purification of Streptomyces isolates
As shown in Table 1, there are 42 isolates of Streptomyces obtained from north Jeddah isolates and from rhizosphere plants of Jeddah gardens, 3 isolates collected from the sea coastal soil in Thual and 2 isolates obtained from green algae.No isolate was obtained the sea shell sample. 5 Streptomyces isolates from rhizosphere plants were obtained from Al Baha.
Table 1. Numbers and locations of Isolated Streptomyces.
|
Samples reagions |
Isolates |
Samples sources |
No. of Samples |
No. of isolates |
|
From Jeddah province |
Soil |
from Plant Rhizosphere plants |
49 |
42 |
|
(Jeddah province) Jeddah city and Thual coast |
Marine samples |
From Red sea shore soil |
4 |
3 |
|
|
|
Green algae |
3 |
2 |
|
|
|
seashells |
3 |
0 |
|
AlBaha |
Soil |
Plants Rhizosphere |
5 |
5 |
Detection of L-asparaginaseactivity (Qualitative assay):
Purified colonies of isolated Streptomyces strains were evaluated for their ability to L-asparaginaseactivity in plates containing M9 agar medium with phenol red. as shown in Table 2 and figure 1, most of the strains isolated had L-asparaginase activity, ranging from 100% to 59.62%. The percentage of the Streptomyces that have L-asparaginase activity from soil were higher than that of sea isolate.
Table 2: L-asparaginaseactivity of Streptomyces isolates (Qualitative assay)
|
Samples regions |
Isolates number |
Positive L-asparaginase activity |
% of Positive of L-asparaginase activity |
|
Soil from Jeddah city |
42 |
22 |
52.38 |
|
Soil from Thual coast |
3 |
3 |
100 |
|
Green Algae |
2 |
2 |
100 |
|
Soil from Al Baha region |
5 |
4 |
80 |
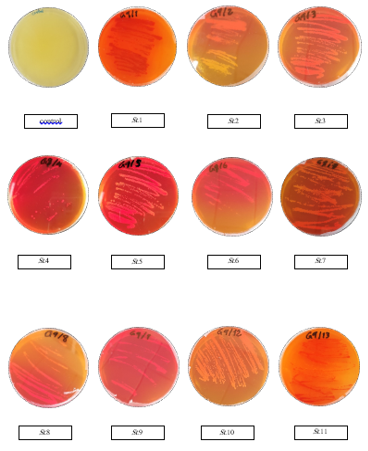
Figure 1: Qualitative Assay to Highest Isolates of Streptomyces Strains
Quantitative assay of L-asparaginase activity by agar well diffusion technique:
Quantitative assay of L-asparaginase in cell-free filtrate performed by diffusion method in a pore on M9 solid media plates, with red phenol, after 24 h of incubation, the measurement of pink area in millimetres were recorded Streptomyces L-asparaginase activity based on the diffused of during 24 h, According to Figure 2, there was no appearance of a pink area on plates number (3, 8, 9), While the pink area was appeared in plates number (1, 2, 4, 5, 6, 7, 10 and 11) and was measured. The number of samples (10, 1 and 11) was recorded as the highest score, and then the number of samples (6, 5, 7, 4 and 2) recorded the lowest score,respectively. According to Fig. 3, which shows the quantitative assay of L-asparaginase by plates,, the different pink zones mean the different of L-asparaginase diffused in solid agar supplemented with L-asparagine compared with control.
Estimation of L-asparaginase activity using nesslerization under Submerged Culture:
The highest L-asparaginase activity of 31 Streptomyces isolates in Cell-Free Filtrate was determined by Nesslerization reaction. The Results in Figure 4 show the different enzyme activity between the isolates showing the highest L-asparaginase activity from the Streptomyces isolates. The highest L-asparaginase activity was from 11 streptomyces isolates.
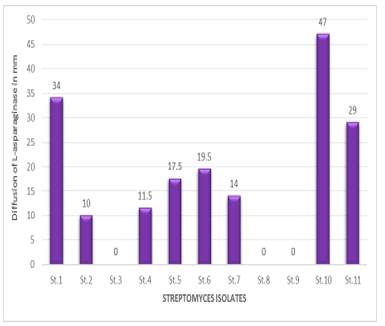
Figure 2 Quantitative assay of L-asparaginase activity by agar well diffusion technique
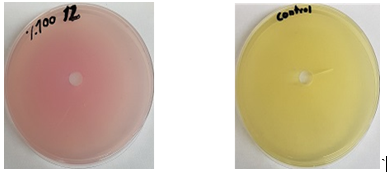
Figure 3: L-asparaginase quantitative in agar, supplemented with L-asparagine.
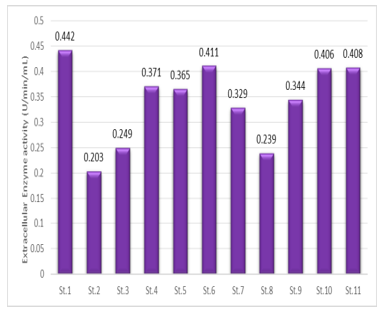
Figure 4: Estimation of L-asparaginase activity of Streptomyces isolates in Submerged Culture
Morphological and Molecular identification
The Results of Table 3 showed some biological studies on the twelve strains in terms of the outward appearance of the strain from the front and back also the pigments that produced in starch nitrate medium. While the internal form or spore chains of the Streptomyces strains
After performing successful PCR as shown in figure 4 at the appropriate length, which is 1.5 kbp, DNA sequences were analyzed using gene bank nucleotide blast alignment tools and showed that the isolates were identified as Streptomyces with similarity percentages 97 - 100% (Table 4). Based on comparative analysis of 16S rDNA sequences of modified and merged isolates selected with the sequence of the closest species recovered by the NCBI BLAST tool, these strains showed taxonomic dependence with isolated strains. The results of phylogenetic relationships of bacterial strains and closely related species were observed using the interactive tree of life (iTOL), ver. 3.4.3 after all sequences were subjected to NCBI blast search tool (Figure 5).
Table 3: Morphological Properties
|
Spore Arrangement |
Diffusible Pigment |
Substrate Mycelium |
Arial Mycelium |
Gram Stain |
No. of isolates |
|
Linear |
Light brown |
Black |
Gray |
+ |
St.1 |
|
Linear |
- |
White to creamy |
White to creamy |
+ |
St.2 |
|
Spiral |
- |
Grayish white |
Gray |
+ |
St.3 |
|
Linear |
- |
White to light gray |
White to light gray |
+ |
St.4 |
|
Spiral |
Yellow |
Orange |
Creamy |
+ |
St.5 |
|
Linear |
- |
Gray to bieage |
grayish brown |
+ |
St.6 |
|
Hook |
- |
Creamy |
White |
+ |
St.7 |
|
Linear |
- |
Red brick |
Red brick |
+ |
St.8 |
|
Spiral |
- |
Creamy |
White |
+ |
St.9 |
|
Spiral |
- |
Red brick |
light grayish red |
+ |
St.10 |
|
Hook |
Yellow |
orange |
white |
+ |
St.11 |
Table 4: Molecular identification of isolates based on 16S rDNA gene.
|
Strains |
Strains |
Gene bank accession numbers |
Identity % |
Coverage % |
|
St.1 |
Streptomyces griseorubens |
MN588164 |
100% |
100% |
|
St. 2 |
Streptomyces alboflavus |
MN588165 |
100% |
100% |
|
St. 3 |
Streptomyces werraensis |
MN588166 |
100% |
100% |
|
St. 4 |
Streptomyces spororaveus |
MN588167 |
99.63% |
100% |
|
St. 5 |
Streptomyces chryseus |
MN588168 |
99.17% |
100% |
|
St. 6 |
Streptomyces globosus |
MN588169 |
99.37% |
99% |
|
St. 7 |
Streptomyces roseolilacinus |
MN588170 |
100% |
100% |
|
St. 8 |
Streptomyces xanthophaeus |
MN588171 |
99.61% |
100% |
|
St.9 |
Streptomyces lavendulae |
MN588172 |
99.60% |
100% |
|
St. 10 |
Streptomyces tuirus |
MN588174 |
100% |
100% |
|
St. 11 |
Streptomyces tendae |
MN588175 |
97.84% |
99% |
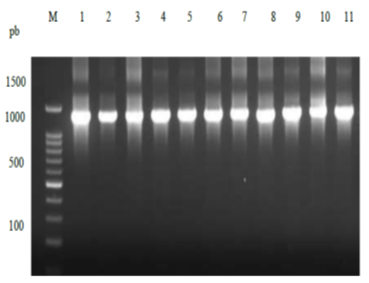
Figure 5: The 1500 bp 16S rDNAampliconagarose gel electrophoresis after the purification compared with DNA Ladder 1kb on a 1 % agarose gel. Lane 1: 1kb DNA leader and Lane 2-12 elven of bacterial isolates.
Figure 6: Neighbor- joining phylogenetic tree based on 16S rRNA sequence analysis and the relationship between Streptomyces and the most closely related bacterial species.
Discussion
Fifty-two different actinomycetes isolates for L-asparaginase, showed 31 positive reactions. These strains were taken for molecular identification. Actinomycetes reactions varies related to the amount of organic matter in plant (Germida et al., 1998). (Tewtrakul and Subhadhirasakul, 2007). root exudates increase the growth of actinomycetes while antimicrobial compounds from the root can reduce the number of soil bacteria and fungi. Actinomycetes are widely distributed in the soil (Velayudham and Murugan, 2012).
Pure colonies from Streptomyces showed approximately 31 of ASNase activity on plate based on the extent of the zone (pink in colour) around the colonies on the Modified M9 plates (Dhanam and Kannan, 2015). Each isolate was subjected to screen for L-asparaginase activity on M9 agar (Baskar and Renganathan, 2012). The formation of a pink zone around the bacteria was a sign of ASNaseproduction (Asselin et al., 1993). Most of the isolated strains have L-asparaginase activity, varied from 100% (isolates that obtained from Soil from the shore of Red Sea coast and Green Algae) to 59.62% (isolated from Soil Al Baha region and Soil from Jeddah province) (Maldonado et al., 2009) showed that actinomycetes are always present in marine sediments, but their numbers are less than soil and these findings are in agreement with the results of this study.
In general, actinomycetes are known to be capable of producing biological activities such as pesticides, herbicides, antibiotics and enzymes including asparaginase (Prapagdee et al., 2008; Boroujeni et al., 2012). Soil samples in Egypt were actinomycete potent antimicrobial compound producing actinomycete were isolates as yellow, grey and white colour series (Atta, 2015). Actinomycetes, especially Streptomyces spp. (Dhevagi and Poorani, 2006; Narayana et al., 2007). Streptomyces Genus is a good source of L-asparaginase compared to bacteria and fungi (Sahu et al., 2007). Actinomycetes revealed to excellent resource for L-asparaginase, as Streptomyces griseus (Narayanaet al., 2007) and in Egypt (EL-sabbagh et al, 2013).
Production and estimation of L-asparaginase anticancer enzyme under SMF by 11 of 31 Streptomyces sp. In the diffusion method plates,there were different pink zones ranged from 10-47 mm, as a result of L-aspraginase hydrolysis activity as explained (Gulati et al., 1997). Our results in accordance with that found by Wakil and Adelegan (2015) with zone ranged from 30-50 cm, and Darwesh et al. (2018) ranged from 30-40 mm. while from 8-13 mm (Devi and Ramanjaneyulu, 2016) and 90mm by (Gulati et al., 1997) in soil bacterial isolates.
More than 75 % of industrial enzymes produced by SMF, which support the use of genetically modified organisms to a greater extent than SSF, and widley SMF is used lack of paraphernalia regarding the production of many enzymes. Highly critical related to metabolism exhibited by microorganisms is different in SSF and SMF, and in a flux of nutrients and efflux of waste needs to carried out based on metabolic parameters (Subramaniyam and Vimala, 2012). Enzyme activity was determined in culture (Dhevagi and Poorani, 2006). Nesslerization reaction was used to estimate the extracellular L-asparaginase activity.
Eleven isolates were selected for further molecular analysis and identification using DNA sequencing of 16S rDNA. To confirm the accuracy, 1500 bpamplicons produced using 16S rDNA universal primers (Edward et al., 1989) were separated on 1 % agarose gels, and then the fragments with the molecular sizes represented the PCR product of 16S rRNA were purified to be ready for sequenced.
DNA sequences were analyzed using nucleotides blast alignment tools of gene bank and showed that the isolates were identified as Streptomyces with similarity percentages 100 % (99% until 97%.). Based on on comparative analysis of 16S rDNA sequences mapped and selected isolates with sequences of nearest type species retrieved by NCBI BLAST tool, these strains showed taxonomic dependence with isolated strains. Isolates were submitted as Streptomyces griseorubenMN588164, Streptomyces alboflavusMN588165, StreptomyceswerraensisMN588166, Streptomyces spororaveusMN588167, Streptomyces chryseusMN588168, Streptomyces globosusMN588169, Streptomyces roseolilacinusMN588170, Streptomyces xanthophaeusMN588171, Streptomyces lavendulaeMN588172, Streptomyces tuirusMN588174 and Streptomyces tendaMN588175.
The 16S rRNA sequence is used to identify Streptomysessp (Khamnaet al., 2009; Deshpande et al., 2014). Even though 16S rRNA gene sequencing is very useful to classifying bacteria, it has a little amount of phylogenetic power (Patel et al., 2001; Bosshard et al., 2006; Mignard and Flandrois, 2006).
Conclusion
Out of 31 L-asparagine strains, only 11 were potential strain for L-asparaginase production. The Organism did not produce any pink zone in control M-9 agar plate incorporated without L-asparagine and the pink zone is only in L-asparaginase production.
References
Alhaithloul, H. A. S. (2019). Environmental and Genetic Diversity of Rangeland Plant Species in Saudi Arabia. World, 8(3), 45-55.
Amena S, Vishalakshi N, Prabhakar M, Dayanand A, & Lingappa K. (2010). Production, purification and characterization of L-asparaginase from Streptomyces gulbargensis. Brazilian journal of Microbiology. 41(1):173-8.
Anisimova, T. Y., Naliukhin, A. N., Hamitowa, S. M., Avdeev, Y. M., & Belozerov, D. A. (2019). Responses of Soil Properties and Crop Productivity to Peat-Fertilizers in Russia. International Journal of Pharmaceutical Research & Allied Sciences, 8(2).
Asselin BL, Whitin JC, Coppola DJ, Rupp IP, Sallan SE, & Cohen HJ. (1993). Comparative pharmacokinetic studies of three asparaginase preparations. Journal of Clinical Oncology. 11(9):1780-6.
Atta HM. (2015). Biochemical studies on antibiotic production from Streptomyces sp.: Taxonomy, fermentation, isolation and biological properties. Journal of Saudi Chemical Society. 19(1):12-22.
Barth PG, Majoie CB, Caan MW, Weterman MA, Kyllerman M, & Smit LM, et al. (2007). Pontine tegmental cap dysplasia: a novel brain malformation with a defect in axonal guidance. Brain. 130(9):2258-66.
Basha NS, Rekha R, Komala M, & Ruby S. (2009). Production of extracellular anti-leukaemic enzyme lasparaginase from marine actinomycetes by solidstate and submerged fermentation: Purification and characterisation. Tropical Journal of Pharmaceutical Research. 8(4).
Baskar G, R& enganathan S. (2012). Optimization of L‐asparaginase production by Aspergillus terreus MTCC 1782 using response surface methodology and artificial neural network‐linked genetic algorithm. Asia‐Pacific Journal of Chemical Engineering. 7(2):212-20.
Boroujeni ME, Das A, Prashanthi K, Suryan S, & Bhattacharya S. (2012). Enzymatic screening and random amplified polymorphic DNA fingerprinting of soil Streptomycetes isolated from Wayanad District in Kerala, India. Journal of Biological Sciences. 12(1):43-50.
Bosshard P, Zbinden R, Abels S, Böddinghaus B, Altwegg M, & Böttger E. (2006). 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory. Journal of clinical microbiology. 44(4):1359-66.
Darwesh OM, Sultan YY, Seif MM, & Marrez DA. (2018). Bio-evaluation of crustacean and fungal nano-chitosan for applying as food ingredient. Toxicology reports. 5:348-56.
Deshpande N, Choubey P, & Agashe M. (2014). Studies on optimization of growth parameters for L-asparaginase production by Streptomyces ginsengisoli. The Scientific World Journal.
Devi A, & Ramanjaneyulu R. (2016). Isolation of L-asparaginase producing microbial strains from soil samples of Telangana and Andhra Pradesh States, India. Int J Curr Microbiol Appl Sci. 5(10):1105-13.
Dhanam J, & Kannan S. (2015). Depiction and screening of L-asparaginase producing actinomycetes isolated from the soil samples of termite mounts. Int J Adv Scientific Technical Res. 1(5):304-10.
Dharmaraj S. (2011). Study of L-asparaginase production by Streptomyces noursei MTCC 10469, isolated from marine sponge Callyspongia diffusa. Iranian journal of biotechnology. 9(2):102-8.
Dhevagi P, & Poorani E. (2006). Isolation and characterization of L-asparaginase from marine actinomycetes. Indian Journal of Biotechnology. 5:514-20.
Duval M, Suciu S, Ferster A, Rialland X, Nelken B, Lutz P, et al. (2002). Comparison of Escherichia coli–asparaginase withErwinia-asparaginase in the treatment of childhood lymphoid malignancies: results of a randomized European Organisation for Research and Treatment of Cancer—Children's Leukemia Group phase 3 trial. Blood. 99(8):2734-9.
Edwards U, Rogall T, Blöcker H, Emde M, & Böttger EC. (1989). Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic acids research. 17(19):7843-53.
El-Sabbagh SM, El-Batanony NH, & Salem TA. (2013). L-Asparaginase produced by Streptomyces strain isolated from Egyptian soil: Purification, characterization and evaluation of its anti-tumor. African Journal of Microbiology Research. 7(50):5677-86.
Germida JJ, Siciliano SD, Renato de Freitas J, & Seib AM. (1998). Diversity of root-associated bacteria associated with field-grown canola (Brassica napus L.) and wheat (Triticumaestivum L.). FEMS Microbiology Ecology. 26(1):43-50.
Graham ML.(2003). Pegaspargase: a review of clinical studies. Advanced drug delivery reviews. 55(10):1293-302.
Gulati R, Saxena R, Gupta R. (1997). A rapid plate assay for screening l‐asparaginase producing micro‐organisms. Letters in applied microbiology. 24(1):23-6.
Gupta N, Mishra S, & Basak U. (2007). Occurrence of Streptomyces aurantiacus in mangroves of Bhitarkanika. Malaysian J Microbiol. 3:7-14.
Jain R, Zaidi K, Verma Y, & Saxena P. (2012). L-asparaginase: A promising enzyme for treatment of acute lymphoblastic leukiemia. People’s Journal of Scientific Research. 5(1):29-35.
Khamna S, Yokota A, & Lumyong S. (2009). L-asparaginase production by actinomycetes isolated from some Thai medicinal plant rhizosphere soils. International Journal of Integrative Biology. 6(1):22-6.
Kozak M, Borek D, Janowski R, & Jaskólski M. (2002). Crystallization and preliminary crystallographic studies of five crystal forms of Escherichia coli L-asparaginase II (Asp90Glu mutant). Acta Crystallographica Section D: Biological Crystallography. 58(1):130-2.
Kumar K, & Verma N. (2012). The various sources and application of L-asparaginase. Asian J Biochem Pharm Res. 2(3):197-205.
Maldonado LA, Fragoso-Yáñez D, Pérez-García A, Rosellón-Druker J, & Quintana ET. (2009). Actinobacterial diversity from marine sediments collected in Mexico. Antonie Van Leeuwenhoek. 95(2):111-20.
Manivasagan P, Venkatesan J, Sivakumar K, & Kim SK. (2013). RETRACTED: marine actinobacterial metabolites: current status and future perspectives.
Mashburn LT. (1964). Tumor inhibitory effect of L-asparaginase from Escherichia coli. Arch Biochem Biophys. 105:450-2.
Mignard S, & Flandrois J-P. (2006). 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. Journal of microbiological methods. 67(3):574-81.
Narayana KJ, Prabhakar P, Vijayalakshmi M, Venkateswarlu Y, & Krishna PS. (2007). Biological activity of phenylpropionic acid isolated from a terrestrial Streptomycetes. Polish journal of microbiology. 56(3):191.
Narta UK, Kanwar SS, & Azmi W. (2007). Pharmacological and clinical evaluation of L-asparaginase in the treatment of leukemia. Critical reviews in oncology/hematology. 61(3):208-21.
Ochi K. (1995). Phylogenetic analysis of mycolic acid-containing wall-chemotype IV actinomycetes and allied taxa by partial sequencing of ribosomal protein AT-L30. International Journal of Systematic and Evolutionary Microbiology. 45(4):653-60.
Pathom-Aree W, Stach JE, Ward AC, Horikoshi K, Bull AT, & Goodfellow M. (2006). Diversity of actinomycetes isolated from Challenger Deep sediment (10,898 m) from the Mariana Trench. Extremophiles. 10(3):181-9.
Patil S, Coutsouvelis J, & Spencer A. (2011). Asparaginase in the management of adult acute lymphoblastic leukaemia: is it used appropriately? Cancer treatment reviews. 37(3):202-7.
Pedreschi F, Kaack K, & Granby K. (2008). The effect of asparaginase on acrylamide formation in French fries. Food chemistry. 109(2):386-92.
Pieters R, Hunger SP, Boos J, Rizzari C, Silverman L, Baruchel A, et al. (2011). L‐asparaginase treatment in acute lymphoblastic leukemia: a focus on Erwinia asparaginase. Cancer. 117(2):238-49.
Prapagdee B, Kuekulvong C, & Mongkolsuk S. (2008). Antifungal potential of extracellular metabolites produced by Streptomyces hygroscopicus against phytopathogenic fungi. International Journal of Biological Sciences. 4(5):330.
Prasad P, Singh T, Bedi S, & Kumari S. (2013). Molecular characterization and phylogenetic analysis of cellulase producing Streptomyces griseorubens (strain st-1) isolated from Indian soil. Jordan Journal of Biological Sciences Short Communication. 6(3).
Radhakrishnan M, Venugopal Gopikrishnan AS, Selvakumar N, & Balagurunathan R. (2013). Characterization and phylogenetic analysis of antituberculous compound producing actinomycete strain D25 isolated from Thar Desert soil, Rajasthan. Bioinformation. 9(1):18.
Rajguru Shubhangi A. & Deshmukh Padma V. (2016). Isolation and screening of L-asparaginase producing bacteria from aquatic and terrestrial habitats in thane district, M.S, India. World jornal of pharmacy and pharmaceutical sciences. (9):1807-1817.
RAMOS CP, Foster G, & Collins MD. (1997). Phylogenetic Analysis of the Genus Actinomyces Based on 16S rRNA Gene Sequences: Description of Arcanobacterium phocae sp. nov., Arcanobacterium bernardiae comb. nov., and Arcanobacterium pyogenes comb. nov. International Journal of Systematic and Evolutionary Microbiology. 47(1):46-53.
Sahu MK, Sivakumar K, Poorani E, Thangaradjou T, & Kannan L. (2007). Studies on L-asparaginase enzyme of actinomycetes isolated from estuarine fishes. Journal of Environmental Biology. 28(2):465.
Savitri NA, & Azmi W. (2003). Microbial L-asparaginase: A potent antitumour enzyme. Indian Journal of Biotechnology. 2:148-94.
Song J, Weon H-Y, Yoon S-H, Park D-S, Go S-J, & Suh J-W. (2001). Phylogenetic diversity of thermophilic actinomycetes and Thermoactinomyces spp. isolated from mushroom composts in Korea based on 16S rRNA gene sequence analysis. FEMS microbiology letters. 202(1):97-102.
Subramaniyam R, & Vimala R. (2012). Solid state and submerged fermentation for the production of bioactive substances: a comparative study. Int J Sci Nat. 3(3):480-6.
Tewtrakul S, & Subhadhirasakul S. (2007). Anti-allergic activity of some selected plants in the Zingiberaceae family. Journal of Ethnopharmacology. 109(3):535-8.
Velayudham S, & Murugan K. (2012). Diversity and antibacterial screening of actinomycetes from Javadi Hill Forest soil, Tamilnadu, India. Journal of Microbiology Research. 2(2):41-6.
Wakil SS, & Adelegan AA. (2015). Screening, production and optimization of L-asparaginase from soil bacteria isolated in Ibadan, South-western Nigeria. Journal of Basic and Applied Sciences. 11:39-51.
Waksman SA, & Lechevalier HA. (1962) The Actinomycetes. Vol. III. Antibiotics of Actinomycetes. The ActinomycetesVol III Antibiotics of Actinomycetes.
ZHANG Q-c, WANG G-h, & YAO H-y. (2007). Phospholipid fatty acid patterns of microbial communities in paddy soil under different fertilizer treatments. Journal of Environmental Sciences.;19(1):55-9.