Diversity Wild Mushrooms in the Community Forest of Na Si Nuan Sub-District, Thailand
Thalisa Yuwa-amornpitak*, Pa-Nag Yeunyaw
Abstract
The aim of this study was to investigate the diversity of edible mushrooms in the community forest of Na Si Nuan sub-district. Twenty-seven species of mushrooms were found, and 24 were edible. Three species were not recorded for eating, and 1 of them was similar to Scleroderma. Mushrooms were identified by their morphology and internal transcribed spacer (ITS) region of ribosomal DNA and then categorized into 13 families. Eight and seven species of mushrooms were placed in Russulaceae and Boletaceae, respectively. Two species of mushrooms were categorized in the families Tricholomataceae and Clavariaceae. Only 1 species was found in each family of Pluteaceae, Amanitaceae, Agaricaceae, Schizophyllaceae, Auriculaceae, Hymeogasteraceae, Diplocystaceae, and Sclerodermataceae. The internal transcribed spacer region of the rDNA sequences of all mushrooms was homologous with the GenBank database, with similarities between 59-100%. Only 9 species from the morphology studies were given the same scientific name as that in the sequence databases: RDEN, REME, AHEM, TCLY, TALB, CMIS, CTA, PHY, and ALPO. The others were the same at the genus level except for SCHI and some Boletus spp. The molecular phylogenetic analysis of 27 mushroom sequences showed high genetics relatedness with reference strains.
Key words: Macrofungi, Mutualistic association, Phylogenetic tree, Internal transcribed spacer (ITS)
Introduction
Functional foods like mushrooms have a wide range of biomolecules with nutritional and medicinal substances (Thaper and Lakshmi, 2017; Mirzaei et al., 2018). Mushrooms are macrofungi (Mansoury, 2019; Al-Zahrani and Bukhari, 2019), and there are approximately 27,000 species widely distributed around the world (Chang and Miles, 2004), with especially high diversity in tropical regions. They are found in a great variety of habitats. Some can grow on rotting logs and animal dung (e.g., saprophyte and parasite), and some mushrooms fruit on the ground beneath or nearby a tree (e.g., ectomycorrhiza). Ectomycorrhizal mushrooms normally live symbiotically with the hairy roots of living plants (Smith and Read, 1997). Currently, more than 100 species of saprophyte mushrooms can be cultivated at the industrial level (Stamets, 1993), such as Pleurotus ostreatus, Pleurotus sajor-caju, and Lentinus squarrosulus. Mycorrhizae represent symbiotic mutualistic relationships between soil fungi and fine plant roots. Ectomycorrhiza (ECM) is a type of mycorrhiza associated with the plant root system, enhancing plant uptake of minerals and drought tolerance. In contrast, the host plant provides photosynthetic products and a place to live to ECM. Dipterocarpaceae contains a group of dominant timber species found in Southeast Asia, including Thailand, and is associated with various types of ECM fungi (Cheevadhanarak and Ratchadawong, 2006). Most of them are Basidiomycetes and Ascomycetes, of them some are edible mushrooms, for example, the most famous and expensive food, truffle (Hall et al., 1998). In addition, most wild edible mushrooms are ectomycorrhizal fungi. Edible mycorrhizal mushrooms have been collected and consumed for centuries.
The classical method for identifying mushrooms is by observing the morphology of the macroscopic (shape and surface of cap, shape and position of stipe, and color), and microscopic structures (structure, shape, size, and color of spores). Though this method takes a long time and is difficult to identify closely related species (Appiah et al., 2017). Therefore, this method needs a well-trained mycologist for identification. The molecular technique is a rapid method, highly specific, and accurate to identify (Aslam et al., 2017). Therefore, this study used morphological structures and molecular techniques to analyze the internal transcript spacer region for identifying and phylogenetic relationships between mushrooms.
Materials and Method
Sample collection
All mushrooms were collected from the local forest of Na Si Nuan sub-district (in a plant genetic conservation project under the Royal Initiative of Her Royal Highness Princess Maha Chakri Sirindhorn at Mahasarakham University), Kantarawichai, Maha Sarakham. The site is located between 16°20’20.5”-16°20’34.6”N and 103°12’44.1”-103°12’45.6”E. The average area covered is approximately 0.065 km2. Small pieces of fresh mushrooms were kept in absolute ethanol and stored at -20°C until use. Complete and fresh sporocarps were identified by morphology using relevant literature (Chandrasrikul et al., 2008; Soithong, 1994; Arora, 1986). The samples were dried at 50°C. Some parts of dried mushrooms were grounded for antioxidant determination (Yuwa-amornpitak et al., 2020).
DNA Extraction and Phylogenetic Tree Construction
The mushroom sample was immersed in absolute ethanol and dried by adsorbing with paper to remove ethanol before DNA extraction with an extraction kit (Vivantis, GF1). The ITS rRNA was amplified by PCR. A diluted genomic DNA template (1 µl) was mixed with 1 µl of each primer (ITS5F: 5’-GGAAGTAAAAGTCGTAACAAGG-3’ and ITS4R: 5’-TCCTCCGCTTATTGATATGC-3’) at a concentration of 20 pmol/µl and master mix with Taq DNA polymerase (Vivantis) for a total volume of 25 µl. Amplification conditions were as follows: denaturation at 94°C for 3 min, 30 cycles of denaturation at 94°C for 40 s, annealing at 55°C for 40 s, and extension at 72°C for 1 min. The final extension was performed at 72°C for 7 min.
The PCR products of the ITS region from each mushroom were sequenced by Sanger Coulson’s method (Sanger et al., 1980) using ABI BigDye (R) Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The sequences were searched for the most similar sequence from the GenBank database with the blastn tool (https://blast.ncbi.nlm.nih.gov/ Blast), as shown in Table 2. The sequences of mushrooms and from the GenBank database were aligned by Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). The alignment of all sequences was examined and adjusted manually by Texpad 8 software. The final alignment was formatted as a MEGA file (MEGA version 6). Neighbor-joining (NJ) analysis was used to generate the distance matrix and produce a tree with 1000 bootstrap replicates. Maximum parsimony (MP) analysis was also conducted by bootstrapping 1000 replicates with MEGA6.0.6. The NJ tree and MP tree are shown in Figures 2 and 3, respectively.
Results and Discussion
Twenty-seven wild mushroom species from the community forest of the Na Si Nuan district were collected and identified (Figure 1). They were studied and identified by morphology. The results are presented in Table 1 with the local name and code. They were placed in 12 families: Russulaceae (8), Boletaceae (7), Tricholomataceae (2), Clavariaceae (2), Plutaceae (2), Lycoperdaceae (1), Schizophyllaceae (1), Auriculaceae (1), Hymeogasteraceae (1), Astracaceae (1), and Sclerodermataceae (1). The code of each species was used to easily remember and represent the local and scientific names. Only in the rainy season (June–October) all mushrooms were found. However, Hed-Poa and Hed-Ra-ngok-luang were found only in the early rainy season (June – July). Hed-Kai-na-kao, Hed-Kai-na-luang, and Hed-Hai-na-keaw were often found in the rainy season, especially when they were abundant at the end of the rainy season. Hed-Pluak-jig and Hed Pluak-tab are termite mushrooms and were also abundant at the end of the rainy season. Termite mushrooms are preferred for eating by local people, including three types of Hed-Kai (Hed-Kai-na-kao, Hed-Kai-na-luang, Hed-Hai-na-keaw). Hed-Kho-na-dang, Hed-Tan-lek, Hed-na-kao, and Boletus species were often found in the rainy season but not in abundance. Hed-Klum-ma, Hed-Sai-duan, Hed-Ta-poo, Hed-Teen-took-kae, Hed-Hoo-noo, and Hed-Pa-ka-lung1 were not abundant. Furthermore, Scleroderma sp. was not abundant and was found only at the end of the rainy season.
From the blastn search tool, the most similar sequence was aligned again with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). The similarity and gap of each sample are shown in Table 2, with the length between 512-810 base pairs. The similarity and gap of each pair were between 59-100% and 0-8.33%, respectively. Most mushroom samples were identified at the genus level. Only 9 samples were fitted to both genus and species levels. These 9 included Russula densifolia, Russula cf. rosaceae, Amanita hemibapha, Termitomyces cylindricus, Termitomyces fuliginosus, Calvatia craniiformis, Clavulina cf. cristata, Phylloporus gajari, and Mycoamaranthus cambodgensis, with percentage similarity (and gap) of 92.98 (0.79), 94.64 (1.17), 96.28(0), 99.70 (0), 82.25(0.49), 99.69(0), 99.69(0), 100(0), and 98.76 (0.15), respectively. Although the similarity of some species was greater than 90%, they were not determined at the species level. These included FLAV, RHET, RALB, SCY, SCHI, AUAR, BOSP, BOL2, BOL3, and ASTA. These results may be because the morphology of each mushroom in the different areas is different and there is no sequence database for those mushrooms in GenBank.
|
|
 |
 |
|
A |
B |
C |
 |
 |
 |
|
D |
E |
F |
 |
 |
 |
|
G |
H |
I |
 |
 |
 |
|
J |
K |
L |
 |
 |
 |
|
M |
N |
O |
 |
 |
 |
|
P |
Q |
R |
 |
 |
 |
|
S |
T |
U |
 |
 |
 |
|
V |
W |
X |
 |
 |
 |
|
Y |
Z |
Ω |
Figure 1. Wild mushrooms collected from the local forest of Na Si Nuan district; mushroom codes: A = RAMY, B = RLIC, C = FLAV, D = RHET, E = RDEN, F = RALB, G = REME, H = LAC, I = AHEM, J = AVAT, K = TCLY, L = TALB, M = CMIS,N = SCY, O = CTA, P = SCHI, Q = AUAR, R = BOSP, S = BOL1, T = BOL2, U = BOL3, V = PHY, W = BOL5, X = BOL6, Y = ALPO, Z = ASTA, Ω = MSU1.
Table 1. Identification of wild mushrooms by morphology, with the local name, code, and scientific name.
|
No. |
Local Name |
Code |
Order |
Family |
Scientific Name |
|
1 |
Hed-Koh-na-dang |
RAMY |
Russulales |
Russulaceae |
Russula emetica |
|
2 |
Hed-Kai-na-kao |
RLIC |
Russulales |
Russulaceae |
Russula delica |
|
3 |
Hed-Kai-na-luang |
FLAV |
Russulales |
Russulaceae |
Russula flavida |
|
4 |
Hed-Kai-na-keaw |
RHET |
Russulales |
Russulaceae |
Russula virescens |
|
5 |
Hed-Tan-lek |
RDEN |
Russulales |
Russulaceae |
Russula densifolia |
|
6 |
Hed-Na-kao |
RALB |
Russulales |
Russulaceae |
Russula alboareolata |
|
7 |
Hed-Dang-ku-lab |
REME |
Russulales |
Russulaceae |
Russula rosacea |
|
8 |
Hed-Nam-noom |
LAC |
Russulales |
Russulaceae |
Lactarius sp. |
|
9 |
Hed-Ra-ngok-luang |
AHEM |
Agaricales |
Pluteaceae |
Amanita hemibapha |
|
10 |
Hed-Sai-duan |
AVAT |
Agaricales |
Pluteaceae |
Amanita vaginata |
|
11 |
Hed-Pluak-jig |
TCLY |
Agaricales |
Tricholomataceae |
Termitomyces clypeatus |
|
12 |
Hed-Pluak-tab |
TALB |
Agaricales |
Tricholomataceae |
Termitomyces fluliginosus |
|
13 |
Hed-Ta-poo |
CMIS |
Agaricales |
Lycoperdaceae |
Calvatia craniformis |
|
14 |
Hed-Pa-ka-lung1 |
SCY |
Agaricales |
Clavariaceae |
Scytinopogon sp. |
|
15 |
Hed-Pa-ka-lung2 |
CTA |
Agaricales |
Clavariaceae |
Clavulina cristata |
|
16 |
Hed-Teen-took-kae |
SCHI |
Agaricales |
Schizophyllaceae |
Schizophyllum commune |
|
17 |
Hed-Hoo-noo |
AUAR |
Auriculariales |
Auriculariaceae |
Auricularia sp. |
|
18 |
Hed-Pung-khom |
BOSP |
Boletales |
Boletaceae |
Boletus sp. |
|
19 |
Hed-Peung-No1 |
BOL1 |
Boletales |
Boletaceae |
Boletus sp. |
|
20 |
Hed-Peung-No.2 |
BOL2 |
Boletales |
Boletaceae |
Boletus sp. |
|
21 |
Hed-Peung-No.3 |
BOL3 |
Boletales |
Boletaceae |
Boletus sp. |
|
22 |
Hed-Peung-No.4 |
PHY |
Boletales |
Boletaceae |
Phylloporus gajari |
|
23 |
Hed-Peung-No.5 |
BOL5 |
Boletales |
Boletaceae |
Boletus sp. |
|
24 |
Hed-Peung-No.6 |
BOL6 |
Boletales |
Boletaceae |
Boletus sp. |
|
25 |
Hed-Klum-ma |
ALPO |
Boletales |
Hymeogasteraceae |
Mycoamaranthus cambodgensis |
|
26 |
Hed-Poa |
ASTA |
Boletales |
Astraceae |
Astraeus hygrometricus |
|
27 |
- |
MSU1 |
Boletales |
Sclerodermata |
Scleroderma sp. |
Table 2: Similarity of ITS sequences of wild mushrooms with the GenBank database.
|
No. |
Local or code name/(code) (Scientific Name)* |
GenBank similar species (Access No.)** |
Similarity, % |
Gap, % |
Alignment base pair |
Access No. |
|
1 |
Hed-Koh-na-dang (RAMY) (Russula emetica) |
Russula lepida (KX655855) |
89.47 |
4.93 |
608 |
MN580109 |
|
2 |
Hed-Kai-na-kao (RLIC) (Russula delica) |
Russula crustosa (EU598193) |
87.31 |
1.83 |
654 |
MN580110 |
|
3 |
Hed-Kai-na-luang (FLAV) (Russula flavida) |
Russula delica (AF345250) |
98.08 |
0 |
600 |
MN580111 |
|
4 |
Hed-Kai-na-keaw (RHET) (Russula virescens) |
Russula aeruginosa (LC008520) |
97.76 |
0.15 |
670 |
MN580112 |
|
5 |
Hed-Tan-lek (RDEN) (Russula densifolia) |
Russula densifolia (AB291763) |
92.98 |
0.79 |
627 |
MN580113 |
|
6 |
Hed-Na-kao (RALB) (Russula alboareolata) |
Unculture Fungus (JN969391) |
94.03 |
0.34 |
586 |
MN580114 |
|
7 |
Hed-Dang-ku-lab (REME) (Russula rosaceae) |
Russula cf. rosaceae (AB459514) |
94.64 |
1.17 |
597 |
MN580115 |
|
8 |
Hed-Nam-noom (LAC) (Lactarius sp.) |
Lactarius picinus (JQ446129) |
83.96 |
1.88 |
636 |
MN580116 |
|
9 |
Hed-Ra-Ngok-Luang AHEM) (Amanita hemibapha) |
Amanita hemibapha (LC068802) |
96.28 |
0 |
512 |
MN580117 |
|
10 |
Hed-Sai-Duan/(AVAT) (Amanita vaginata) |
Amanita sp. (AB854646) |
76.17 |
3.06 |
554 |
MN580118 |
|
11 |
Hed-Pluak-Jig/(TCLY) (Termitomyces clypeatus) |
Termitomyces cylindricus (LC068786) |
99.70 |
0 |
673 |
MN580119 |
|
12 |
Hed-Pluak-Tab/(TALB) (Termitomyces fuliginosus) |
Termitomyces fuliginosus (KX781739) |
82.25 |
0.49 |
648 |
MN580120 |
|
13 |
Hed-Ta-poo (CMIS) (Calvatia craniiformis) |
Calvatia craniiformis (MH916598) |
99.69 |
0 |
649 |
MN580121 |
|
14 |
Hed-Pa-ka-lang 1 (SCY) (Scytinopogon sp.) |
Scytinopogon sp. (KT804576) |
95.71 |
0.49 |
607 |
MN580122 |
|
15 |
Hed-Pa-ka-lang 2(CTA) (Clavulina cistata) |
Clavulina cf. cristata (AB459513) |
99.69 |
0 |
664 |
MN580123 |
|
16 |
Hed-Teen-took-kae (SCHI) (Schizophyllum commune) |
Hohenbuehelia grisea (MF150036) |
94.51 |
0.81 |
614 |
MN580124 |
Table 2 (continued)
|
No. |
Local or code name/(code) (Scientific Name)* |
GenBank similar species (Access No.)** |
Similarity, % |
Gap, % |
Alignment base pair |
Access No. |
|
17 |
Hed-Hoo-noo (AUAR) (Auricularia sp.) |
Auricularia cornea (KX022016) |
98.22 |
0.178 |
561 |
MN580125 |
|
18 |
Hed-Pung-khom (BOSP) (Boletus sp.) |
Boletus griseipurpureus (KF442409) |
94.70 |
1.06 |
661 |
MN580126 |
|
19 |
Hed-Peung-No.1 (BOL1) (Boletus sp.) |
Tylopilus plubeoviolaceoides (KM975490) |
77.05 |
9.11 |
558 |
MN580127 |
|
20 |
Hed-Peung-No.2 (BOL2) (Boletus sp.) |
Indoporus shorea (NR161157) |
95.05 |
0.09 |
526 |
MN580127 |
|
21 |
Hed-Peung-No.3 (BOL3) Boletus sp. |
Sepedonium chrysospermum (KT946851) |
96.17 |
0.19 |
522 |
MN580129 |
|
22 |
Hed-Peung-No.4 (PHY) (Phylloporus gajari) |
Phylloporus gajari (NR155649) |
100 |
0 |
588 |
MN580130 |
|
23 |
Hed-Peung-No.5 (BOL5) (Boletus sp.) |
Boletus fraternus (MH211928) |
72.32 |
1.53 |
655 |
MN580131 |
|
24 |
Hed-Peung-No.6 (BOL6) (Boletus sp.) |
Boletus sp. MES235 (FJ480441) |
79.11 |
4.15 |
747 |
MN580132 |
|
25 |
Hed-Klum-ma (ALPO) (Mycoamaranthus cambodgensis) |
Mycoamaranthus cambodgensis (LC068799) |
98.76 |
0.15 |
647 |
MN203105 |
|
26 |
Hed-Poa (ASTA) (Astraeus hygrometricus) |
Astraeus asiaticus (EU718089) |
96.58 |
0.28 |
702 |
MN203104 |
|
27 |
(MSU1) (Scleroderma sp.) |
Scleroderma sp. (MG833748) |
59.45 |
8.33 |
624 |
MN203106 |
*Scientific name by morphological identification
Molecular phylogenetic trees from neighbor-joining and maximum parsimony methods were analyzed by MEGA 6.0.6 and are shown in Figures 2 and 3, respectively. Both trees showed the same placement of each species with homologous ITS sequence from GenBank database and percentage confident placing with the 1000 replicate bootstraps. The NJ and MP tree consisted of 3 clades (A, B, and C). The Russulales, Agaricales and Aphyllophoreales orders were closely related and grouped in clade A. However, the results also showed that the order Boletales was divergent, and it separated into 2 clades, B and C. However, the two clades were sisters.
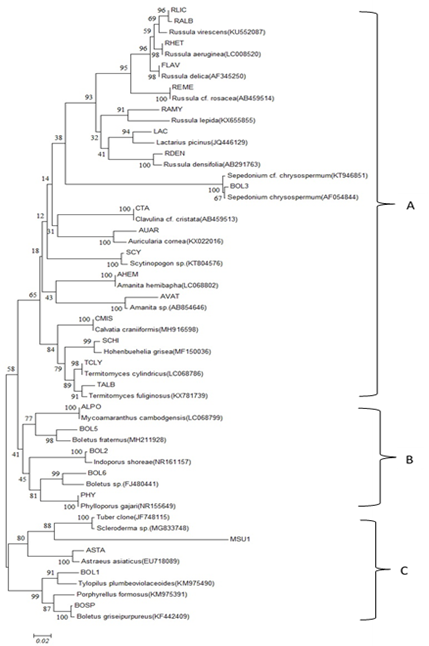
Figure 2. Neighbor-joining tree generated from the ITS regions of 27 wild mushrooms (code). Numbers on nodes indicate the bootstrap percentage of 1000 replicates.
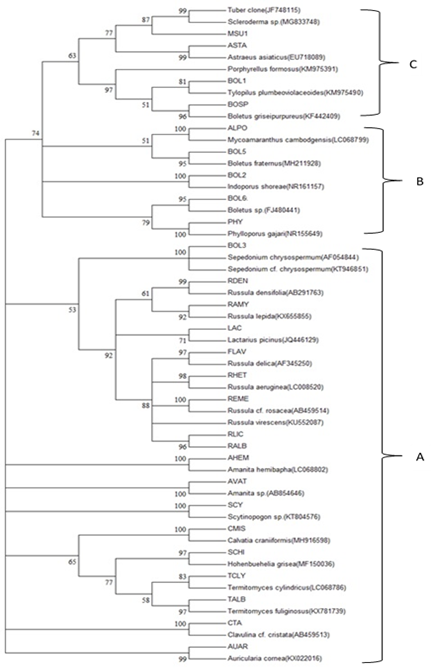
Figure 3. The consensus tree of the maximum parsimony trees generated by the ITS regions of 27 wild mushrooms (code). The number on each node indicates the bootstrap percentage of 1000 replicates.
In addition, the tree showed 100 percentages from bootstrapping, which revealed they were very closely related. They included ALPO, BOL2, BOL3, PHY, REME, AHEM, AVAT, SCY, CMIS, and CTA, which were highly confident for identifying Mycoamaranthus cambodgensis, Indoporus shorea, Sepedonium chrysospermum, Phylloporus gajari, Russula cf. rosaceae, Amanita hemibapha, Amanita sp., Scytinopogon sp., Calvatia craniiformis, and Clavulina cf. cristata, respectively. This study indicated that the local forest of the Na Si Nuan district is a small area but still has a diverse selection of wild edible mushrooms. Dipterocarp forest is still the dominant timber tree that produces various types of mushrooms for mushroom hunters.
There are color differences in the mushroom cap in Russula specie of Hed-kai group (white, yellow, and green). They were identified as Hed-kai-na-kao (Russula delica, RLIC), Hed-kai-na-luang (R. flavida, FLAV), and Hed-kai-na-keaw (R. virescences, RHET), respectively. However, the ITS sequences of those mushrooms were blasted against sequences from GenBank database; the results showed that the first, second and the third species were similar to Russula crustose (Accession no. EU598193), Russula delica (AF345250), and Russula aeruginosa (LC008520) with the similarity of 87.31, 98.08, and 97.76%, respectively. However, they were not matched with morphological identification. Furthermore, more results showed that the identification by morphology was not matched with sequence alignment despite high similarity (more than 90%) as shown in Table 2, for example, Hed-Poa (ASTA), Hed-teen took-kae (SCHI), and Hed-Pluak-Jig (TCLY).
In this study, fruit bodies of 27 species of wild mushrooms were collected. The relationship between them was analyzed and the molecular phylogenetic tree was reconstructed. It was found that most of the species (18 species) were grouped in clade A. They were grill and jelly mushrooms of the order Russulales, Agaricales, and Auricularles. Clade B and C were very closely related. They were placed in the order Boletales (9 species). Some species of mushrooms that were found in this study were distributed in other areas; for example, Amanita hemibapha was also found in Phuphan National Park, Thailand (Wongchalee and Pukahute, 2012) and Eastern Chota Nagpur Plateau of West Bengal in India (Das et al., 2013). Schizophyllum commune is reported from many countries, such as Nigeria (Adeniyi et al., 2018), China (Appiah et al., 2017), India (Ao et al., 2016), and Thailand (in this study). Astraeus hygrometricus was also collected by Das and colleagues (Das et al., 2013). However, molecular identification of mushrooms with 100% homology of gene sequence are difficult, due to the difference in the gene sequence by the difference ecological areas where mushrooms are present (Olusegun, 2014).
Conclusions
Although the local forest of Na Si Nuan district is a small area, but still there are diverse wild edible mushrooms. Dry Dipterocarp forest is still the dominant timber tree that produced various types of mushrooms for local mushroom hunters.
Acknowledgments
We appreciate the plant genetic conservation project under the Royal Initiative of Her Royal Highness Princess Maha Chakri Sirindhorn. This work was supported by two grants from the Faculty of Technology and Mahasarakham University (project number 6105056).
References
Adeniyi, M., Titilawo, Y., Oluduro, A., Odeyemi, O., Nakin, M., & Okoh, A. I. (2018). Molecular identification of some wild Nigerian mushrooms using internal transcribed spacer: polymerase chain reaction. AMB Express, 8(1), 1-9.
Al-Zahrani, S. H. M. & Bukhari, K. A. (2019). Genetics Identification And Detection Of L-Methioninase Gene In A Pseudomonas Isolate. Pharmacophore, 10(6), 93-98.
Ao, T., Seb, J., Ajungla, T., & Deb, C. R. (2016). Diversity of wild mushrooms in Nagaland, India. Open Journal of Forestry, 6(5), 404-419.
Appiah, T., Agyare, C., & Luo, Y. (2017). Molecular Identification of Some Ghanaian Mushrooms Using Internal Transcribed Spacer Regions. Bio 6:1-5.
Arora, D. Mushrooms demystified: A comprehensive guide to the fleshy fungi. 1986. Berkeley: Ten Speed Press, 959p.
Aslam, S., Tahir, A., Aslam, M. F., Alam, M. W., Shedayi, A. A., & Sadia, S. (2017). Recent advances in molecular techniques for the identification of phytopathogenic fungi–a mini review. Journal of Plant Interactions, 12(1), 493-504.
Chandrasrikul, A., Suwanarit, P., Sangwanit, U., Morinaga, T., Nishizawa, Y., & Murakami, Y. (2008). Diversity of mushrooms and macrofungi in Thailand. Kasetsart University, Bangkok.
Cheevadhanarak, S., & Ratchadawong, S. (2006). Diversity of ectomycorrhizal fungi on Dipterocarpaceae in Thailand. Journal of Biological Sciences, 6(6), 1059-1064.
Das SK, Mandal A, Datta AK, Gupta S, Paul R, Saha A, Sengupta S, & Dubey PK. (2013). Nucleotide sequencing and identification of some wild mushrooms. The Scientific World Journal. Article ID 403191
Hall, I. R., Zambonelli, A., & Primavera, F. (1998). Ectomycorrhizal fungi with edible fruiting bodies 3. Tuber magnatum, Tuberaceae. Economic botany, 52(2), 192-200.
Mansoury M. M. S. (2019). Evidence-Based Therapeutic Activity of Pomegranate And Its Active Constituent Ellagic Acid. Pharmacophore, 10(1), 30-36.
Miles, P. G., & Chang, S. T. (2004). Mushrooms: cultivation, nutritional value, medicinal effect, and environmental impact. CRC press.
Mirzaei, A., Torbatinezhad, N. & Razavi, S. E. (2018). Effect of pleurotous eryngii mushroom-treated wheat straw on the function of afshari fattened lambs. Journal of Advanced Pharmacy Education & Research, 8(S2), 189-191.
Olusegun, O. V. (2014). Molecular identification of Trametes species collected from Ondo and Oyo States, Nigeria. Jordan Journal of Biological Sciences, 147(1572), 1-5.
Sanger, F., Coulson, A., Barrell, B. G., Smith, A. J. H., & Roe, B. A. (1980). Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. Journal of molecular biology, 143(2), 161-178.
Smith, S. E., & Read, D. J. (1997). Mycorrhizal symbiosis, 2nd ed. Academic Press, California.
Soithong K (1994). Mushrooms and macrofungi in Thailand. Siritham Offset Publishers Ltd. Ubonratchathani, Thailand.
Stamets P (1993) Growing gourmet and medicinal mushrooms, Ten Speed Press, Berekely, California.
Thaper, S. & Lakshmi, T. (2017). Effects of mushroom on dental caries. Journal of Advanced Pharmacy Education & Research, 7(3), 197-199.
Wongchalee, P., & Pukahute, C. (2012). Diversity of mushrooms in dry dipterocarp forest at Phuphan national park, Sakon Nakhon Province. Natural Science, 4(12), 1153-1160.
Yuwa-amornpitak T, Butkhup L, Yeunyaw P (2020) Amino acids and antioxidant activities of extracts from wild edible mushrooms from a community forest in the Nasrinual District, Maha Sarakham, Thailand. Food Science and Technology, Ahead of Print.