Synthesis in microwave, pharmacological evaluation, molecular docking and ADME studies of Schiff bases of Diclofenac targeting COX-2
Yasir M. Kadhim*, Souad J. Lafta and Monther F. Mahdi
Abstract
Objective: To synthesize and preliminary pharmacological evaluation of new analogs of several new Schiff bases of diclofenac by investigating their interactions with COX-2 by docking and defining certain associations between their structures to improve selectivity towards COX-2 and to decrease side effects. Methods: Several new diclofenac Schiff bases were designed, prepared, and tested as potential COX-2 inhibitors. These new compounds were tested by molecular docking using genetic optimization for ligand docking suite for evaluated of their in vivo anti-inflammatory activity and COX-2 selectivity. Results: Because of their hydrogen bonding interaction with main amino acids in COX isozymes Arg121, Tyr356, and Ser120, all compounds evaluated in molecular docking exhibited important activities compared with diclofenac and 4PH9 as comparison drugs. The results of the ADME showed that all synthesized compounds absorbed from GIT while all compounds except (Yr 05 h-j) followed the Lipinski law. Conclusion: The production of the designed compounds has been managed successfully, the anti-inflammatory evaluation of the end products suggests that the new Schiff bases derivatives have strengthened their anti-inflammatory action, docking studies have shown that the preliminary analysis of anti-inflammatory activity has shown that all compounds (Yr 04, Yr 05 a-j) have strong anti-inflammatory properties, excluding Yr 03.
Key words: Diclofenac, Schiff base, docking, ADME, GOLD, Lipinski rule, microwave irradiations
Introduction
|
|
Diclofenac is a proven and widely used non-steroidal anti-inflammatory drug (NSAID) of the phenylacetic acid course. As with most NSAIDs, diclofenac exerts anti-inflammatory, analgesic, and antipyretic actions via inhibition of cyclooxygenases (COXs). The first created of Diclofenac was achieved by Alfred Sallmann and Rudolf Pfister in 1973 and since then the drug has been in clinical use for the treatment of rheumatoid arthritis, osteoarthritis, ankylosing spondylitis, gout attacks, postoperative pain, dysmenorrhea, and various ocular conditions (Davies and Anderson, 1997).
Besides its use in pathological processes related to inflammation, diclofenac and its chemical derivatives have been thoroughly examined for a varied range of pharmacological effects (Rossoni et al., 2008; Frantzias et al., 2012; Končič et al., 2009; Rojo et al., 2009; Johnsen et al., 2004; Wittine et al., 2009; Moody et al., 2010; Zhang et al., 2009). Several scientific communications report the broad-spectrum antibacterial activity of diclofenac in vitro and in vivo against isolates of various gram-positive and gram-negative bacteria that were either sensitive or resistant to antibiotics (Muñoz-Criado et al., 1996; Dastidar et al., 2000; Mazumdar et al., 2006; Mazumdar et al., 2009). The drug was reported to be highly bactericidal and exerts its action by inhibiting the DNA synthesis of bacteria (Dastidar et al., 2000; Mazumdar et al., 2009). Likewise, several small molecules bearing the 2-[(2,6-dichlorophenyl)amino]benzyl unit were shown to possess antibacterial potential (Sriram et al., 2006; Patel and Patel, 2011).
Antiviral activity of diclofenac was first studied by Gordon et al. in 1998. These researchers investigated topical NSAIDs direct pharmacological effects on adenoviral replication in vitro and only diclofenac appeared to have an inhibitory effect against different adenovirus serotypes (Gordon et al., 1998).
In a recent patent, the inventors claimed that diclofenac or its pharmaceutically acceptable salt inhibits the activity of herpes viruses, including herpes simplex virus type 1 (HSV-1), herpes simplex virus type 2 (HSV-2), and varicella-zoster virus. The drug was found to reduce the lesion size, lesion number, and viral titer when topically administered to herpesvirus lesions in animals (Lee and Shieh, 2011).
In a more recent study, some NSAIDs, antioxidants, and peroxisome proliferator-activated receptor-gamma agonists were tried for their capacity to interfere with rotavirus ECwt (wild type) infectivity in ICR mice. The work has to indicate that the treatment of rotavirus infected mice with diclofenac led to the reduced infection of villus cells (Guerrero et al., 2013).
The function and morphology of the COX enzymes
The prostaglandins are lipid autacoids resulting from arachidonic acid and are synthesized via the cyclooxygenase pathway. Two linked isoforms of the cyclooxygenase enzyme have been defined. Cyclooxygenase-1 (COX-1) is accountable for the physiological creation of prostanoids while the cyclooxygenase-2 (COX-2) reasons the raised creation of prostanoids that happen at the site of disease and inflammation. COX-1 is defined as a "house-keeping enzyme" that controls ordinary cellular routes like gastric cytoprotection, vascular homeostasis, platelet aggregation, and kidney function. COX-2 is constitutively communicated in some tissues such as the brain, kidney, and bone, and its appearance at other sites is improved through states of inflammation. The two enzymes portion sixty-present homology in their amino acid sequence. However the complicated conformations for the substrate-binding places and catalytic areas are somewhat changed, for example, COX-2 has a greater and more elastic substrate station than COX-1 has, and also, COX-2 has a greater space at the site where inhibition bind and this structural change between COX-1 and COX-2 has allowed the development of COX-2 selective inhibiter (Picot et al., 1994).
Fig. 1 illustrates a diagram depiction of the amino acid structural variances between the substrate-binding channels of COX-1 and COX-2 that permitted the design of selective inhibitors. It is strong that the amino acid residues Val434, Arg513, and Val523 form a side compact in COX-2 that is absent in COX-1.
These findings resulted in that: (a) The non-selective inhibitors have access to the binding channels of both isoforms, while, (b) The more voluminous residues in COX-1, that are Ile434, His513, and Ile532, obstruct access of the bulky side chains of the COX-2 inhibitors (Grosser et al., 2006).
Studies (Liou, 2010), have shown that glucocorticoids can suppress the creation of proteins involved in inflammation (resulting in their role as anti-inflammatory compounds). Aside from that, glucocorticoids further suppress inflammation by activating a group of enzymes known as lipocortins. Lipocortins have been found to inhibit or decrease the activity of phospholipase A2, a key enzyme that participates in the release of arachidonic acid from the cell membrane, where it is usually incorporated into. When the cell attacked by foreign substances, arachidonic acid is released from the cell membrane and is converted into substances such as prostaglandins which mediate inflammation. Free arachidonic acid is being used in the production of inflammatory prostaglandins by the COX-2 isozyme. The release of arachidonic acids requires the activation of the enzyme phospholipase A2. As mentioned earlier the lipocortins inhibit the phospholipase A2 activity. By activating lipocortins, glucocorticoids cause the inhibition of phospholipase A2, thereby inhibiting the release of arachidonic acid and consequently prostaglandin synthesis in the cell and therefore participate in their effect as significant anti-inflammatory agents. Because lower amounts of inflammatory prostaglandins are synthesized, inflammation is suppressed and damage caused by chronic inflammation is decreased.
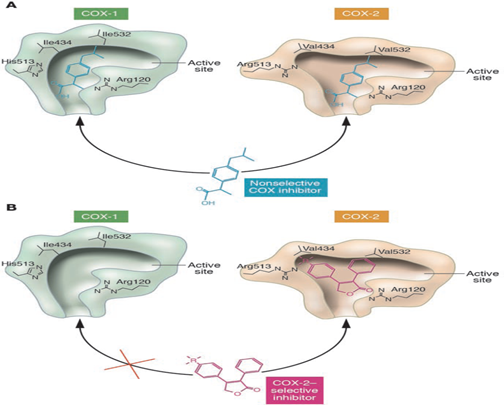
Fig. (1): The schematic depiction of the amino acid structural differences between the substrate-binding channels of COX-1 and COX-2 that allowed the design of selective inhibitors (Grosser et al., 2006).
Heterocyclic Compounds containing an azomethine group (- CH=N-), called as Schiff bases. The first synthesis of imine was done by Hugo Schiff in 1864 (Hussain et al., 2014). The Schiff base is formed by reacting compounds having active carbonyl groups with primary amines in presence acid (Saxena, 2013). Schiff bases possess a wide variety of biological activities such as antimicrobial activity (Al-Shemary et al., 2016), antileishmanial (Al Zoubi, 2013), anti-inflammatory (Tantaru et al., 2013), anti-HIV (Patel et al., 2012), Anticonvulsant (Singh and Kumar, 2016), anticancer (Arulmurugan et al., 2010), antifungal (Kailas et al., 2016) and anti-proliferative (Su et al., 2015). Schiff bases derivatives are important in the medical field.
Microwave chemistry is the science of applying microwave radiation to chemical reactions (Microwaves in Organic Synthesis, 2018; de la Hoz et al., 2005; Strauss and Trainor, 1995; Kidwai, 2001; Kappe et al., 2012). Microwaves act as high-frequency electric fields and will generally heat any material containing mobile electric charges, such as polar molecules in a solvent or conducting ions in a solid. Polar solvents are heated as their component molecules are forced to rotate with the field and lose energy in collisions. Semiconducting and conducting samples heat when ions or electrons within them form an electric current and energy is lost due to the electrical resistance of the material. Microwave heating in the laboratory began to gain wide acceptance following papers in 1986 (Gedye et al., 1986), although the use of microwave heating in chemical modification can be traced back to the 1950s. Although occasionally known by such acronyms as MAOS (Microwave-Assisted Organic Synthesis) (Pizzetti et al., 2012), MEC (Microwave-Enhanced Chemistry), or MORE synthesis (Microwave-organic Reaction Enhancement), these acronyms have had little acceptance outside a small number of groups.
Materials and Methods
General
All reagents and anhydrous solvents were of an annular type and generally used as received from the commercial suppliers (Merck, Germany, Reidel-De Haen, Germany, Sigma-Aldrich, Germany and BDH, England).
Experimental Part
Synthesis of [2-(2,6-dichloroanilino)phenyl]acetic acid (Yr 02)
Diclofenac sodium (2g, 6.28mmol), was dissolved in 50ml of warm water with stirring for 10 minutes, then 2N HCl (3.1ml, 6.28mmol) was added, followed by addition of excess cold water (100ml), the acid was precipitated then filtered, dried and used in the following step without further purification (Banerjee and Amidon, 1981).
IR spectra (KBr, ʋmax, cm-1): 3323 (N‑H), 3066 (O‑H), 1689 (C=O), 1303 (C‑O), 740 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 3.72 (s, 2H, CH2), 6.29 (s, 1H, NH), 6.85–7.53 (m, 7H, ArH), 12.73 (s, 1H, OH).
Synthesis of methyl [2‑(2,6‑dichloroanilino)phenyl]acetate (Yr 03)
The diclofenac methyl ester was prepared as per the procedure described previously in the literature (Ballini and Carott, 1983). These steroidal carboxylic acids were methylated using Potassium Carbonate as a base in refluxing acetone.
Yield: 98% , mp: 97.5°C.
IR spectra (KBr, ʋmax, cm-1): 3352 (N‑H), 3024 (C‑H), 1737 (C=O), 1294 (C‑O), 750 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 3.66 (s, 3H, O-CH3), 3.82 (s, 2H, CH2), 6.26 (s, 1H, NH), 6.84–7.55 (m, 7H, ArH).
Synthesis of 2‑[2‑(2,6‑dichloroanilino) phenyl]acetohydrazide (Yr 04)
Compound (Yr 03, 0.01 mol) and hydrazine hydrate (0.02 mmol) were refluxed in absolute ethanol (50 ml) for 24 h (examined by thin-layer chromatography [TLC]).
Compound synthesis (Yr 04) was performed as stated in previous literature (Palkar et al., 2014). The mixture was concentrated, cooled, and placed in ice water. So isolated white amorphous solid was washed, dried, and recrystallized from ethanol and water to provide compound (Yr 04).
Yield: 99 % , mp 158°C.
IR spectra (KBr,ʋmax, cm-1): 3348(NH2), 3327 (N‑H), 3030 (C‑H), 1635 (C=O), 742 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 3.36 (s, 2H, CH2), 4.35 (s, 2H, NH2), 6.30 (s, 1H, NH), 6.85–7.53 (m, 7H, ArH), 9.51 (s, 1H, CO-NH).
General method for the production of 2‑(2‑(2,6‑dichloroanilino)phenyl)‑N’‑ ethylideneaceto hydrazide derivatives (Yr 05a‑j)
A finely ground mixture of Compound (Yr 04, 0.01 mmol) was added in the irradiation tube, the appropriately substituted aromatic aldehyde (0.01 mmol) was added with 2 drops from glacial acetic acid and the reaction mix was irradiated in microwave synthesis reactor (450w) for a suitable time. Then, the reaction combination was cool to the room temperature, filtered, and the Schiff base is collected. The product was recrystallized using absolute ethanol as per the earlier report.
2-[2-(2,6-dichloroanilino)phenyl]-N'-[(E)-(pyridin-4-yl)methylidene]acetohydrazide (Yr 05a)
Yield: 71 % , mp 228°C.
IR spectra (KBr,ʋmax, cm-1): 3317 (N‑H), 3028 (C‑H), 1647 (C=O), 744 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 4.17 (s, 2H, CH2), 6.29 (s, 1H, NH), 6.84–8.65 (m, 11H, ArH), 11.90 (s, 1H, N=CH), 12.10 (s, 1H, CO-NH).
2-[2-(2,6-dichloroanilino)phenyl]-N'-[(1E)-1-(pyridin-4-yl)ethylidene]acetohydrazide (Yr 05b)
Yield: 75 % , mp 241-243°C.
IR spectra (KBr,ʋmax, cm-1): 3275 (N‑H), 3024 (C‑H), 1660 (C=O), 759 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 2.30 (s, 3H, CH3), 4.21 (s, 2H, CH2), 6.30 (s, 1H, NH), 6.84–8.62 (m, 11H, ArH), 10.99 (s, 1H, CO-NH).
2-[2-(2,6-dichloroanilino)phenyl]-N'-[(1E)-1-(pyridin-3-yl)ethylidene]acetohydrazide (Yr 05c)
Yield: 73 % , mp 250°C.
IR spectra (KBr,ʋmax, cm-1): 3288 (N‑H), 3057 (C‑H), 1668 (C=O), 752 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 2.34 (s, 3H, CH3), 4.19 (s, 2H, CH2), 6.28 (s, 1H, NH), 6.84–9.03 (m, 11H, ArH), 11.00 (s, 1H, CO-NH).
2-[2-(2,6-dichloroanilino)phenyl]-N'-[(E)-(furan-2-yl)methylidene]acetohydrazide (Yr 05d)
Yield: 89 % , mp 267°C.
IR spectra (KBr,ʋmax, cm-1): 3288 (N‑H), 3022 (C‑H), 1643 (C=O), 746 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 4.06 (s, 2H, CH2), 6.26 (s, 1H, NH), 6.32–8.13 (m, 10H, ArH), 11.55 (s, 1H, N=CH), 11.74 (s, 1H, CO-NH).
2-[2-(2,6-dichloroanilino)phenyl]-N'-[(1E)-1-(furan-2-yl)ethylidene]acetohydrazide (Yr 05e)
Yield: 88 % , mp 281°C.
IR spectra (KBr,ʋmax, cm-1): 3248 (N‑H), 3068 (C‑H), 1660 (C=O), 746 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 2.20 (s, 3H, CH3), 4.09 (s, 2H, CH2), 6.21 (s, 1H, NH), 6.31–7.81 (m, 10H, ArH), 10.82 (s, 1H, CO-NH).
2-[2-(2,6-dichloroanilino)phenyl]-N'-[(E)-(thiophen-2-yl)methylidene]acetohydrazide (Yr 05f)
Yield: 70 % , mp 278°C.
IR spectra (KBr,ʋmax, cm-1): 3273 (N‑H), 3021 (C‑H), 1643 (C=O), 746 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 4.06 (s, 2H, CH2), 6.28 (s, 1H, NH), 6.83–8.24 (m, 10H, ArH), 11.59 (s, 1H, N=CH), 11.74 (s, 1H, CO-NH).
2-[2-(2,6-dichloroanilino)phenyl]-N'-[(1E)-1-(thiophen-2-yl)ethylidene]acetohydrazide (Yr 05g)
Yield: 79 % , mp 287°C.
IR spectra (KBr,ʋmax, cm-1): 3269 (N‑H), 3026 (C‑H), 1662 (C=O), 746 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 2.31 (s, 3H, CH3), 4.10 (s, 2H, CH2), 6.28 (s, 1H, NH), 6.84–7.85 (m, 10H, ArH), 10.90 (s, 1H, CO-NH).
N'-[(E)-(4-chlorophenyl)methylidene]-2-[2-(2,6-dichloroanilino)phenyl]acetohydrazide (Yr 05h)
Yield: 75 % , mp 227°C.
IR spectra (KBr,ʋmax, cm-1): 3340 (N‑H), 3068 (C‑H), 1656 (C=O), 750 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 4.12 (s, 2H, CH2), 6.28 (s, 1H, NH), 6.32–8.17 (m, 11H, ArH), 11.59 (s, 1H, N=CH), 11.78 (s, 1H, CO-NH).
2-[2-(2,6-dichloroanilino)phenyl]-N'-[(E)-(4-dimethylanilino)methylidene]acetohydrazide (Yr 05i)
Yield: 76 % , mp 250°C.
IR spectra (KBr,ʋmax, cm-1): 3317 (N‑H), 3028 (C‑H), 1647 (C=O), 744 (C‑Cl).
1H‑NMR (DMSO, δ, ppm): 3.32 (s, 6H, CH3), 4.10 (s, 2H, CH2), 6.30 (s, 1H, NH), 6.86–8.26 (m, 11H, ArH), 11.32 (s, 1H, N=CH), 11.49 (s, 1H, CO-NH).
2-[2-(2,6-dichloroanilino)phenyl]-N'-[(E)-(4-nitrophenyl)methylidene]acetohydrazide (Yr 05j)
Yield: 78 % , mp 242°C.
IR spectra (KBr,ʋmax, cm-1): 3362 (N‑H), 3080 (C‑H), 1658 (C=O), 750 (C‑Cl).
Computational Method
The computational method approved in this work is outlined in Fig. 2. A full licensed CCDC genetic optimization for ligand docking (GOLD) Suite (v. 5.7.3) was used to achieve the molecular docking studies for the compounds. CCDC Hermes visualizer software (v. 1.10.3) was used to envisage: the protein, ligands, interactions of hydrogen bonding, short contacts, and length of bonds calculation. The chemical structures of our ligands were drawn using ChemDraw Professional software (v. 16.0) Fig. 3.
The pharmacokinetic profile, i.e., adsorption, distribution, metabolism, excretion (ADME) of the synthesized compounds was expected with the assistance of the Swiss ADME server. (Daina et al., 2017)

Fig. (2) Outline of computational procedure.

Fig. (3) A full licensed CCDC genetic optimization for ligand docking (GOLD)
ADME procedures
All ligands were drawn by ChemSketch (v. 14), converted to SMILE name by Swiss ADME tool which predicts the physicochemical descriptors and pharmacokinetic properties. BOILED-EGG was used to compute the lipophilicity and polarity of the small molecule. (Friedman, 1951)
Preparation of ligands and protein receptor
The crystal structures of the enzyme COX 2 [PDB ID: 4PH9] were downloaded from the Protein Data Bank (PDB) and their missing atoms were added with the assistance of Swiss PDB Viewer (v. 3.7). The crystal structures of our downloaded protein were prepared by eliminating all water molecules and by the addition of hydrogen atoms to get a right ionization and tautomeric states of amino acid residues. Chem3D (v. 16.0) was used to minimize the energy for our synthesized ligands by applying the MM2 force field.
Docking procedures
The full license version of GOLD (v. 5.7.3) was used for molecular docking. (Jones et al., 1997; Jones et al., 1995) The Hermes visualizer software in the GOLD Suite was used to set up the receptors for the docking process additionally. The binding site used to dock GOLD has been identified as all the protein residues inside the 10 Å of the source ligands that occur in the complexes of the downloaded protein structure. Two COX-2 proteins were downloaded from the PDB website (1PXX and 4PH9) to dock the whole process. (Huang and Zou, 2007) As a result, 4PH9 was selected to dock the compounds.
CCDC Superstar was used to assessing the cavity and the active site. The protein reference ligand has been used for determining the active site radius (10 Å). ChemScore kinase was used as a template for the configuration. For the scoring feature ChemPiecewise linear potential (CHEMPLP) was utilized. The scores of all parameters used during the docking process remained the default, and all solutions are scored according to the fitness function of CHEMPLP. According to CHEMPLP, the steric adjunct between protein and ligand is determined while the distance and angle-dependent hydrogen are considered. The results of docking, i.e., the binding mode, docked pose, and binding free energy was studied to evaluate the interaction between the amino acid residues of the proteins COX-2 and our synthesized ligands.
Molecular Modeling
Genetic optimization for ligand docking is a “genetic algorithm for docking flexible ligands into protein binding sites” (Webb and Griswold, 1984). GOLD has been broadly verified and has shown superb rendering for pose prediction and excellent results for virtual screening (Palm et al., 1997). It is provided as a part of the GOLD Suite, which contains additional software components, Hermes, Mercury, Isostar and Conquest, and GoldMine, etc.
Ligands and protein-energy minimization will repair distorted geometries by transferring atoms to release internal restrictions. After the minimization of the energy, the geometry is fixed which means a minimum of energy has been reached.
To predict the selectivity and binding energies of the synthesized compounds for COX-1 and COX-2, docking studies were performed with the help of GOLD Suite software to study the molecular interactions involved in between active binding sites of the protein target and the synthesized compounds (Table 1).
Table 1: The binding energies for derivatives and reference NSAIDs docked with COX-2
|
Compounds |
COX-2 binding energy (PLP Fitness) |
No. of Amino acids included in H-bonding |
Amino acids included in H-bonding |
no. of bonding |
Length of bonding |
|
|
4PH9 |
66.32 |
3 |
ARG 121 |
1 |
3.047 |
|
|
ARG 121 |
1 |
3.007 |
||||
|
TYR 356 |
1 |
2.761 |
||||
|
YR 05 i |
93.22 |
3 |
TYR 356 |
2 |
3.036 |
2.597 |
|
ARG 121 |
1 |
3.025 |
||||
|
YR 05 a |
84.93 |
2 |
ARG 121 |
1 |
3.034 |
|
|
TYR 356 |
1 |
2.771 |
||||
|
YR 05 f |
84.06 |
3 |
ARG 121 |
1 |
2.950 |
|
|
ARG 121 |
1 |
2.877 |
||||
|
TYR 356 |
1 |
2.542 |
||||
|
YR 05 h |
83.39 |
3 |
ARG 121 |
1 |
3.059 |
|
|
ARG 121 |
1 |
2.952 |
||||
|
TYR 356 |
1 |
2.498 |
||||
|
YR 05 d |
79.39 |
3 |
ARG 121 |
1 |
2.916 |
|
|
ARG 121 |
1 |
2.854 |
||||
|
TYR 356 |
1 |
2.456 |
||||
|
YR 05 j |
77.92 |
1 |
SER 531 |
1 |
2.979 |
|
|
YR 05 g |
77.47 |
2 |
ARG 121 |
1 |
2.984 |
|
|
TYR 356 |
1 |
2.892 |
||||
|
YR 05 c |
75.93 |
5 |
SER 120 |
1 |
3.006 |
|
|
ARG 121 |
2 |
3.019 |
2.738 |
|||
|
ARG 121 |
1 |
3.012 |
||||
|
TYR 356 |
1 |
2.967 |
||||
|
YR 05 e |
72.42 |
3 |
ARG 121 |
1 |
2.976 |
|
|
ARG 121 |
1 |
2.870 |
||||
|
TYR 356 |
1 |
2.747 |
||||
|
YR 05 b |
70.61 |
2 |
ARG 121 |
1 |
2.923 |
|
|
TYR 356 |
1 |
2.681 |
||||
|
YR 04 |
68.34 |
4 |
HIS 90 |
1 |
3.080 |
|
|
LEU 353 |
1 |
2.770 |
||||
|
SER 354 |
2 |
2.738 |
2.462 |
|||
|
YR 03 |
62.66 |
1 |
TYR 356 |
1 |
2.733 |
|
The distance of short contacts and hydrogen bonding between a specific protein atom and our synthesized ligands is calculated by GOLD and all bond length below 3 Å. (Verdonk et al., 2003) The short contacts are described as other interacting forces like van der Waals, electrostatic, steric, p – p stacking, dipole-dipole, and others.
All the synthesized compounds having promising docking results with COXs, fitted in the COX-2 active site as shown in Fig. 4–16
 |
 |
|
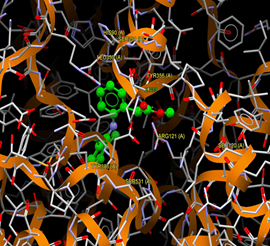
Fig. (4) : Short contacts interaction profile for the compound reference 4PH9. The interaction between compound (4PH9) and amino acid residues [4Ph9: Ball-and-stick style, residues of amino acid in the capped form]. |
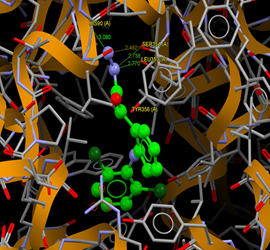
Fig. (5) : Short contacts interaction profile for the compound Yr 03. The interaction between compound (Yr 03) and amino acid residues [Yr 03: Ball-and-stick style, residues of amino acid in the capped form]. |
 |
 |
|
Fig. (6) : Short contacts interaction profile for the compound Yr 04. The interaction between compound (Yr 04) and amino acid residues [Yr 04: Ball-and-stick style, residues of amino acid in the capped form]. |
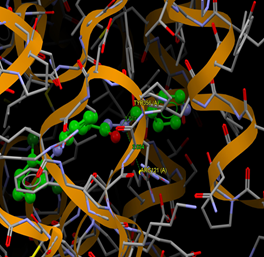
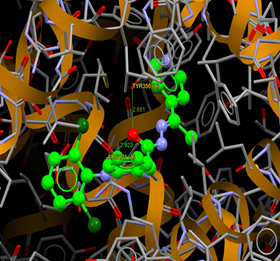
Fig. (7) : Short contacts interaction profile for the compound Yr 05 a. The interaction between compound (Yr 05 a) and amino acid residues [Yr 05 a: Ball-and-stick style, residues of amino acid in the capped form]. |
 |
 |
|
Fig. (8) : Short contacts interaction profile for the compound Yr 05 b. The interaction between compound (Yr 05 b) and amino acid residues [Yr 05 b: Ball-and-stick style, residues of amino acid in the capped form]. |
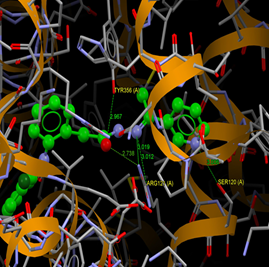
Fig. (9) : Short contacts interaction profile for the compound Yr 05 c. The interaction between compound (Yr 05 c) and amino acid residues [Yr 05 c: Ball-and-stick style, residues of amino acid in the capped form]. |
 |
 |
|
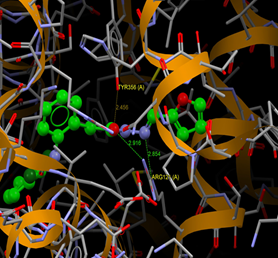
Fig. (10) : Short contacts interaction profile for the compound Yr 05 d. The interaction between compound (Yr 05 d) and amino acid residues [Yr 05 d: Ball-and-stick style, residues of amino acid in the capped form]. |
Fig. (11) : Short contacts interaction profile for the compound Yr 05 e. The interaction between compound (Yr 05 e) and amino acid residues [Yr 05 e: Ball-and-stick style, residues of amino acid in the capped form]. |
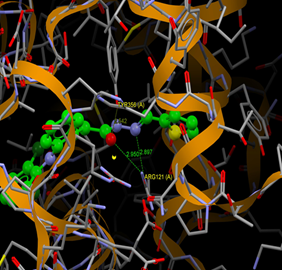 |
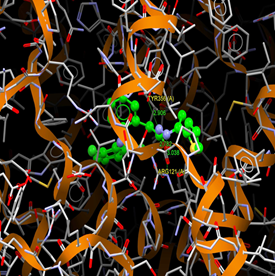 |
|
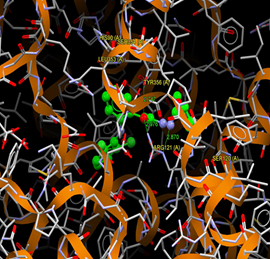
Fig. (12) : Short contacts interaction profile for the compound Yr 05 f. The interaction between compound (Yr 05 f) and amino acid residues [Yr 05 f: Ball-and-stick style, residues of amino acid in the capped form]. |
Fig. (13) : Short contacts interaction profile for the compound Yr 05 g. The interaction between compound (Yr 05 g) and amino acid residues [Yr 05 g: Ball-and-stick style, residues of amino acid in the capped form]. |
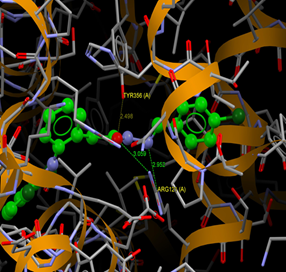 |
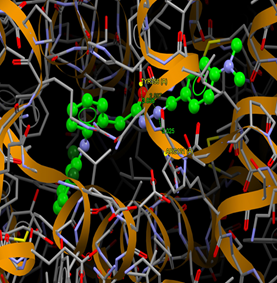 |
|
Fig. (14) : Short contacts interaction profile for the compound Yr 05 h. The interaction between compound (Yr 05 h) and amino acid residues [Yr 05 h: Ball-and-stick style, residues of amino acid in the capped form]. |
Fig. (15) : Short contacts interaction profile for the compound Yr 05 i. The interaction between compound (Yr 05 i) and amino acid residues [Yr 05 i: Ball-and-stick style, residues of amino acid in the capped form]. |
|
|
|
|
Fig. (16) : Short contacts interaction profile for the compound Yr 05 j. The interaction between compound (Yr 05 j) and amino acid residues [Yr 05 j: Ball-and-stick style, residues of amino acid in the capped form].
|
|
Compounds Yr 05 (a-i) (Figs. 7–15) show H-bond interactions with Arg121 and Tyr356 and these two amino acids exist in the binding with five approved NSAIDs (Ibuprofen, Naproxen, Indomethacin, Flurbiprofen, and Des-methylflurbiprofen). Compound (Yr 05 j) (Fig. 16) has H-bond with Ser531 which is the binding site of diclofenac, lumiracoxib, tolfenamic acid. Compound (Yr 03) (Fig. 5) has one H-bond with Tyr355 like in aspirin.
ADME Studies
The ADME properties profile of our created compounds was studied by the Swiss ADME server to detect the safer and potential drug candidate(s) to filter out the compounds which are most likely to fail in the subsequent stages of drug development due to unfavorable ADME properties.
We assessed all synthesized compounds' ADME method.
Also, we measured the topological polar surface area (TPSA), as this is another important property related to the bioavailability of drugs. Thus, passively absorbed molecules with a TPSA > 140 Å are thought to have low oral bioavailability. (Suralkar et al., 2008) Our findings indicate that all produced compounds have TPSA below 140, which is within the range of 38–99 and that all ligands have a bioavailability of 0.55, indicating that all ligands enter the systemic circulation.
Compounds Yr (02-04) and Yr 05 (a-g) fulfilled Lipinski rule. Also, it fulfilled the topological descriptors and fingerprints of molecular drug-likeness structure keys as LogP and LogS.
The GI absorption score is a measure of the extent of absorption of a molecule from the intestine following oral administration. The absorption could be excellent if the result were high. In this study, the GI absorption of all compounds was high predicting them to be well absorbed from the intestine.
The ADME properties profiles for the created compounds are shown in (Table 2).
Table 2: ADME properties profile of the synthesized compounds
|
Comps. |
Formula |
M.Wt (g/mol) |
H-bond acceptors |
H-bond donors |
MR |
TPSA Ų |
GI Abs. |
BBB permeant |
Lipinski violations |
|
Yr 02 |
C14H11Cl2NO2 |
296.15 |
2 |
2 |
77.55 |
49.33 |
High |
Yes |
0 |
|
Yr 03 |
C15H13Cl2NO2 |
310.17 |
2 |
1 |
81.87 |
38.33 |
High |
Yes |
0 |
|
Yr 04 |
C14H13Cl2N3O |
310.18 |
2 |
3 |
81.49 |
67.15 |
High |
Yes |
0 |
|
Yr 05 a |
C20H16Cl2N4O |
399.28 |
3 |
2 |
109.55 |
66.38 |
High |
Yes |
0 |
|
Yr 05 b |
C21H18Cl2N4O |
413.30 |
3 |
2 |
114.36 |
66.38 |
High |
No |
0 |
|
Yr 05 c |
C21H18Cl2N4O |
413.30 |
3 |
2 |
114.36 |
66.38 |
High |
No |
0 |
|
Yr 05 d |
C19H15Cl2N3O2 |
388.25 |
3 |
2 |
104.03 |
66.63 |
High |
Yes |
0 |
|
Yr 05 e |
C20H17Cl2N3O2 |
402.28 |
3 |
2 |
108.83 |
66.63 |
High |
No |
0 |
|
Yr 05 f |
C19H15Cl2N3OS |
404.31 |
2 |
2 |
109.64 |
81.73 |
High |
No |
0 |
|
Yr 05 g |
C20H17Cl2N3OS |
418.34 |
2 |
2 |
114.44 |
81.73 |
High |
No |
0 |
|
Yr 05 h |
C21H16Cl3N3O |
432.73 |
2 |
2 |
116.77 |
53.49 |
High |
No |
1 |
|
Yr 05 i |
C23H22Cl2N4O |
441.36 |
2 |
2 |
125.97 |
56.73 |
High |
Yes |
1 |
|
Yr 05 j |
C21H16Cl2N4O3 |
443.28 |
4 |
2 |
120.58 |
99.31 |
High |
No |
1 |
Conclusion
References
Al-Shemary, R. K., Al-khazraji, A. M., & Niseaf, A. N. (2016). Preparation, spectroscopic study of Schiff base ligand complexes with some metal ions and evaluation of antibacterial activity. The Pharma Innovation, 5(1, Part B), 81.
Arulmurugan, S., Kavitha, H. P., & Venkatraman, B. R. (2010). Biological activities of Schiff base and its complexes: a review. Rasayan J Chem, 3(3), 385-410.
Ballini, R., & Carott, A. (1983). A simple and mild procedure for the preparation of Bile acid methyl esters. Synthetic Communications, 13(14), 1197-1201.
Banerjee, P. K., & Amidon, G. L. (1981). Physicochemical property modification strategies based on enzyme substrate specificities I: Rationale, synthesis, and pharmaceutical properties of aspirin derivatives. Journal of pharmaceutical sciences, 70(12), 1299-1303.
Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific reports, 7, 42717.
Dastidar, S. G., Ganguly, K., Chaudhuri, K., & Chakrabarty, A. N. (2000). The anti-bacterial action of diclofenac shown by inhibition of DNA synthesis. International journal of antimicrobial agents, 14(3), 249-251.
Davies, N. M., & Anderson, K. E. (1997). Clinical pharmacokinetics of diclofenac. Clinical pharmacokinetics, 33(3), 184-213.
de la Hoz, A., Diaz-Ortiz, A., & Moreno, A. (2005). Microwaves in organic synthesis. Thermal and non-thermal microwave effects. Chemical Society Reviews, 34(2), 164-178.
Frantzias, J., Logan, J. G., Mollat, P., Sparatore, A., Del Soldato, P., Ralston, S. H., & Idris, A. I. (2012). Hydrogen sulphide‐releasing diclofenac derivatives inhibit breast cancer‐induced osteoclastogenesis in vitro and prevent osteolysis ex vivo. British journal of pharmacology, 165(6), 1914-1925.
Friedman, H. L. (1951). Influence of isosteric replacements upon biological activity. Nasnrs, 206, 295-358.
Gedye, R., Smith, F., Westaway, K., Ali, H., & Baldisera, L. (1986). The use of microwave ovens for rapid organic synthesis. Tetrahedron letters, 27(3), 279-282.
Gordon, Y. J., Araullo-Cruz, T., & Romanowski, E. G. (1998). The effects of topical nonsteroidal anti-inflammatory drugs on adenoviral replication. Archives of Ophthalmology, 116(7), 900-905.
Grosser, T., Fries, S., & FitzGerald, G. A. (2006). Biological basis for the cardiovascular consequences of COX-2 inhibition: therapeutic challenges and opportunities. The Journal of clinical investigation, 116(1), 4-15.
Guerrero, C. A., Pardo, P., Rodriguez, V., Guerrero, R., & Acosta, O. (2013). Inhibition of rotavirus ECwt infection in ICR suckling mice by N-acetylcysteine, peroxisome proliferator-activated receptor gamma agonists and cyclooxygenase-2 inhibitors. Memórias do Instituto Oswaldo Cruz, 108(6), 741-754.
Huang, S. Y., & Zou, X. (2007). Ensemble docking of multiple protein structures: considering protein structural variations in molecular docking. Proteins: Structure, Function, and Bioinformatics, 66(2), 399-421.
Hussain, Z., Yousif, E., Ahmed, A., & Altaie, A. (2014). Synthesis and characterization of Schiff's bases of sulfamethoxazole. Organic and medicinal chemistry letters, 4(1), 1.
Johnsen, J. I., Lindskog, M., Ponthan, F., Pettersen, I., Elfman, L., Orrego, A., ... & Kogner, P. (2004). Cyclooxygenase-2 is expressed in neuroblastoma, and nonsteroidal anti-inflammatory drugs induce apoptosis and inhibit tumor growth in vivo. Cancer research, 64(20), 7210-7215.
Jones, G., Willett, P., & Glen, R. C. (1995). Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. Journal of molecular biology, 245(1), 43-53.
Jones, G., Willett, P., Glen, R. C., Leach, A. R., & Taylor, R. (1997). Development and validation of a genetic algorithm for flexible docking. Journal of molecular biology, 267(3), 727-748.
Kailas, H. K., Sheetal, P. J., Anita, P., & Apoorva, H. (2016). Four synthesis methods of Schiff base ligands and preparation of their metal complex with IR and antimicrobial investigation. World Journal of Pharmacy and Pharmaceutical Sciences, 5(2), 1055-1063.
Kappe, C. O., Stadler, A., & Dallinger, D. (2012). Microwaves in organic and medicinal chemistry (Vol. 52). John Wiley & Sons.
Kidwai, M. (2001). Dry media reactions. Pure and Applied Chemistry, 73(1), 147-151.
Končič, M., Rajič, Z., Petrič, N., & Zorc, B. (2009). Antioxidant activity of NSAID hydroxamic acids. Acta pharmaceutica, 59(2), 235-242.
Lee, F., & Shieh, H. L. (2011). US Patent 2011/0092597.
Liou S, (2010). "Inflammation", Hopes, Huntington's outreach project for education at Stanford, Stanford, CA 94305, United States.
Mazumdar, K., Dastidar, S. G., Park, J. H., & Dutta, N. K. (2009). The anti-inflammatory non-antibiotic helper compound diclofenac: an antibacterial drug target. European journal of clinical microbiology & infectious diseases, 28(8), 881.
Mazumdar, K., Dutta, N. K., Dastidar, S. G., Motohashi, N., & Shirataki, Y. (2006). Diclofenac in the management of E. coli urinary tract infections. in vivo, 20(5), 613-619.
Microwaves in Organic Synthesis. Organic Chemistry Portal. Retrieved 23 October 2018.
Moody, T. W., Switzer, C., Santana-Flores, W., Ridnour, L. A., Berna, M., Thill, M., ... & Roberts, D. D. (2010). Dithiolethione modified valproate and diclofenac increase E-cadherin expression and decrease proliferation of non-small cell lung cancer cells. Lung Cancer, 68(2), 154-160.
Muñoz-Criado, S., Muñoz-Bellido, X. L., & Garcia-Rodriguez, J. A. (1996). In vitro activity of nonsteroidal antiinflammatory agents, phenotiazines, and antidepressants againstBrucella species. European Journal of Clinical Microbiology and Infectious Diseases, 15(5), 418-420.
Palkar, M. B., Singhai, A. S., Ronad, P. M., Vishwanathswamy, A. H. M., Boreddy, T. S., Veerapur, V. P., ... & Karpoormath, R. (2014). Synthesis, pharmacological screening and in silico studies of new class of Diclofenac analogues as a promising anti-inflammatory agents. Bioorganic & medicinal chemistry, 22(10), 2855-2866.
Palm, K., Stenberg, P., Luthman, K., & Artursson, P. (1997). Polar molecular surface properties predict the intestinal absorption of drugs in humans. Pharmaceutical research, 14(5), 568-571.
Patel, N. B., & Patel, J. C. (2011). Synthesis and antimicrobial activities of 2-azetidinyl-4-quinazolinone derivatives of diclofenac analogue. Medicinal Chemistry Research, 20(4), 511-521.
Patel, R. N., Patel, P. V., Desai, K. R., Purohit, P. Y., Nimavat, K. S., & Vyas, K. B. (2012). Synthesis of new heterocyclic schiff base, thiazolidinone and azetidinone compounds and their antibacterial activity and anti-hiv activities. Heterocyclic Letters, 2(1), 99-105.
Picot, D., Loll, P. J., & Garavito, R. M. (1994). The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1. Nature, 367(6460), 243-249.
Pizzetti, M., Petricci, E., & Tecnologico, C. (2012). Heterogeneous catalysis under microwave heating. La Chimica & L’Industria, 12, 78-81.
Rojo, C., Alvarez-Figueroa, M. J., Soto, M., Canete, A., Pessoa-Mahana, D., & López-Alarcón, C. (2009). Scavenging activity of diclofenac: interaction with ABTS radical cation and peroxyl radicals. Journal of the Chilean Chemical Society, 54(1), 58-62.
Rossoni, G., Sparatore, A., Tazzari, V., Manfredi, B., Soldato, P. D., & Berti, F. (2008). The hydrogen sulphide‐releasing derivative of diclofenac protects against ischaemia–reperfusion injury in the isolated rabbit heart. British journal of pharmacology, 153(1), 100-109.
Saxena, A. (2013). Synthesis and characterization of schiff base salicylaldehyde and thiohydrazones and its metal complexes. Advances in Applied Science Research, 4(4), 152-54.
Singh, I., & Kumar, A. (2016), Synthesis, Characterization, And Antifungal Activity of Schiff Base And Its Metal Complexes, European Journal of Pharmaceutical And Medical Research, 3(6), 363-365.
Sriram, D., Yogeeswari, P., & Devakaram, R. V. (2006). Synthesis, in vitro and in vivo antimycobacterial activities of diclofenac acid hydrazones and amides. Bioorganic & medicinal chemistry, 14(9), 3113-3118.
Strauss, C. R., & Trainor, R. W. (1995). Developments in microwave-assisted organic chemistry. Australian Journal of Chemistry, 48(10), 1665-1692.
Su, R., Lü, L., Zheng, S., Jin, Y., & An, S. (2015). Synthesis and characterization of novel azo-containing or azoxy-containing Schiff bases and their antiproliferative and cytotoxic activities. Chemical Research in Chinese Universities, 31(1), 60-64.
Suralkar, A. A., Sarda, P. S., Ghaisas, M. M., Thakare, V. N., & Deshpande, A. D. (2008). In-vivo animal models for evaluation of anti-inflammatory activity. Latest Rev, 6(2).
Tantaru, G., Nechifor, M., & Profire, L. (2013). Synthesis and biological evaluation of some new Schiff bases and their Cu (II) and Mg (ІІ) complexes. African Journal of Pharmacy and Pharmacology, 7(20), 1225-1230.
Verdonk, M. L., Cole, J. C., Hartshorn, M. J., Murray, C. W., & Taylor, R. D. (2003). Improved protein–ligand docking using GOLD. Proteins: Structure, Function, and Bioinformatics, 52(4), 609-623.
Webb, E. F., & Griswold, D. E. (1984). Microprocessor-assisted plethysmograph for the measurment of mouse paw volume. Journal of pharmacological methods, 12(2), 149-153.
Wittine, K., Benci, K., Rajić, Z., Zorc, B., Kralj, M., Marjanović, M., ... & Balzarini, J. (2009). The novel phosphoramidate derivatives of NSAID 3-hydroxypropylamides: Synthesis, cytostatic and antiviral activity evaluations. European journal of medicinal chemistry, 44(1), 143-151.
Zhang, J., Wang, J., Wu, H., He, Y., Zhu, G., Cui, X., & Tang, L. (2009). Design, synthesis and insulin-sensitizing activity of indomethacin and diclofenac derivatives. Bioorganic & medicinal chemistry letters, 19(12), 3324-3327.

